Figure 2.
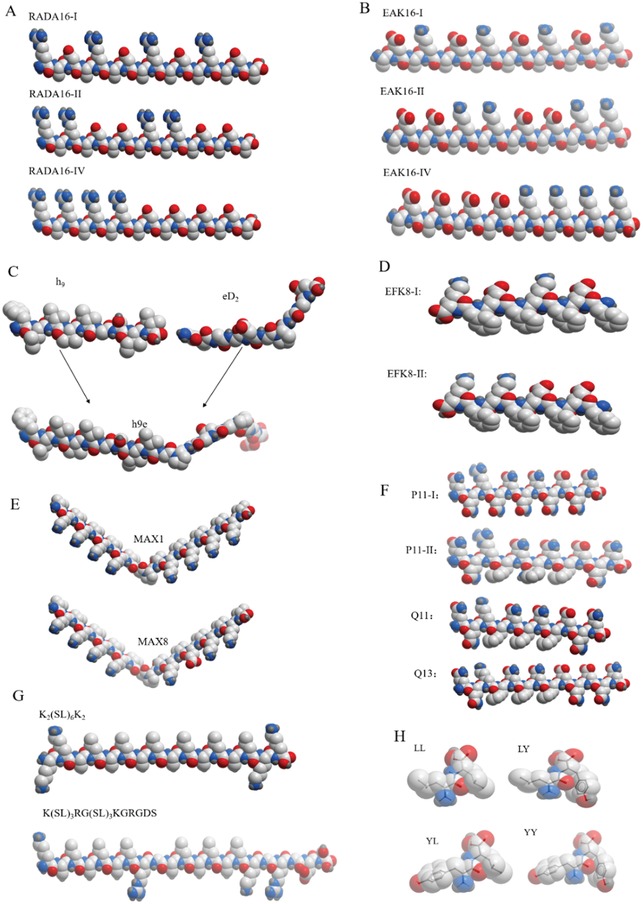
Molecular models of designer self‐assembling peptides currently available in biomedical applications (some other designer peptides are not enclosed here). A) RADA16 modulus peptide shows alternating charged amino acid residues on hydrophilic surface side. The hydrophobic amino acid residues are rationally localized to another β‐sheet side. B) EAK16 modulus peptide shows completely similar molecular building block arrangement with RADA16 modulus peptide except that arginine and aspartic acid residues substitute lysine and glutamate in sequence for salt‐facilitated scaffold formation, since the stable β‐sheet formed is important not only for nanofiber architecture but also for hydrogel formation.[ 215 ] C) h9e peptide molecular model consists of eD2 and h9 peptide fragments. D) Molecular model of EFK8 peptide (EFK8‐I and EFK8‐II). E) Molecular models of MAX1 and MAX8 peptides, where only one amino acid difference occurs in sequence. F) Molecular models of glutamate‐rich self‐assembling peptides (P11‐I, P11‐II, Q11, and bQ13). G) The multidomain peptides are tethered by the well‐known three amino acid cell adhesion motif (RGD) to promote this variant compatible for cell culture.[ 123b ] H) Molecular models of dipeptides (LL, LY, YL, and YY) obtained by atomistic molecular simulations. All short peptide models are produced using the ICM‐Browser software package (MolSoft LLC, San Diego, CA, USA).
