Figure 6: Gene set enrichment analysis of genes upregulated in the GFP, DP, YFP, and DN groups compared to published data sets.
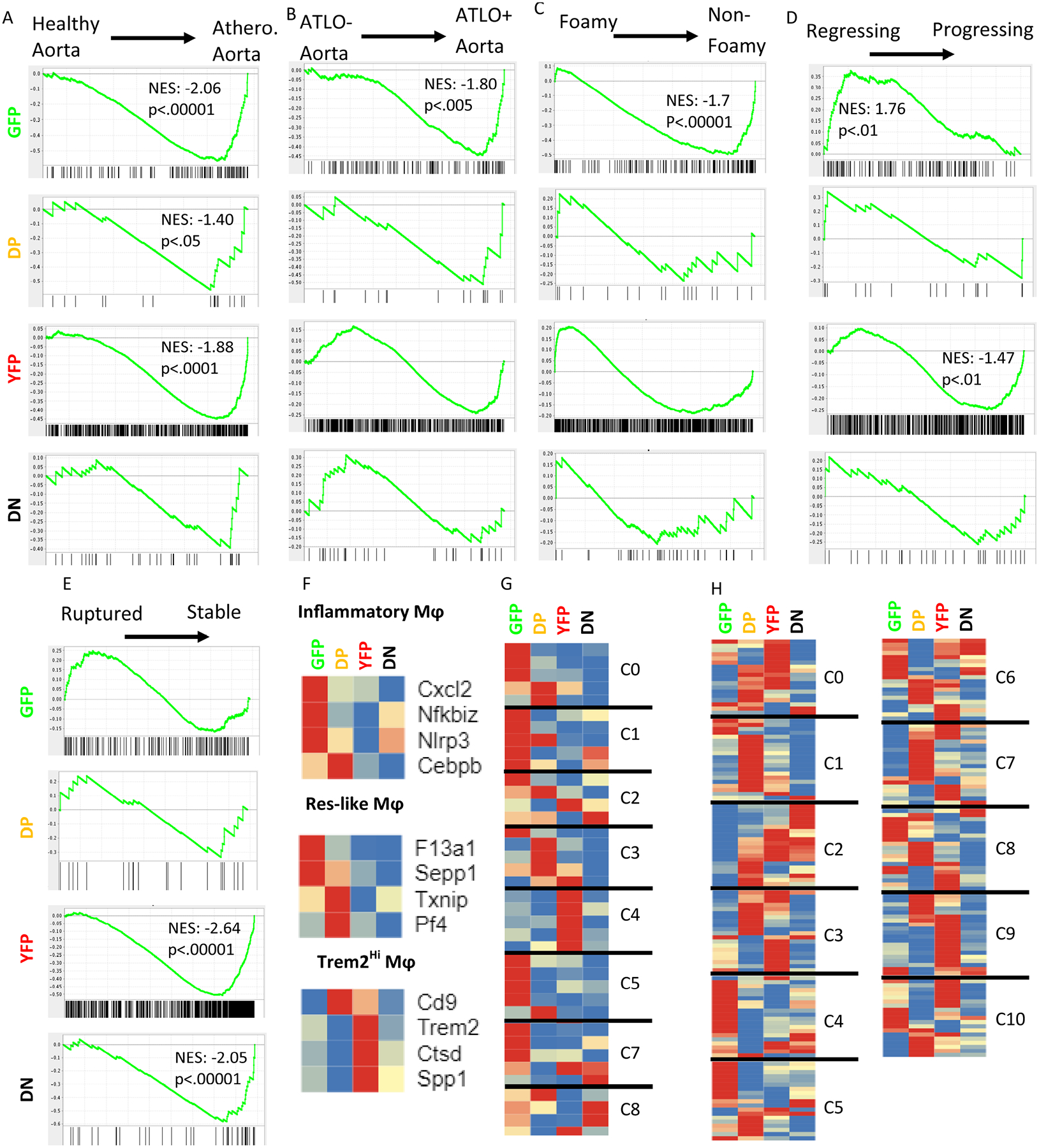
A) Comparison to whole vessel wall transcriptomes from healthy and atherosclerotic mouse aortas (GSE10000)34. B) Comparison to transcriptomes of atherosclerotic aortas with and without ATLO (GSE10000)34. C) Comparison to transcriptomes of atherosclerotic foamy or non-foamy macrophages (GSE11623914) D) Comparison to transcriptomes from regressing and progressing plaques based on a surgical model (GSE24819)35. All p-values listed are False Discovery Rate q-values. E) Comparison to transcriptomes of macrophages from ruptured or stable human atheromatous plaques derived from carotid endarterectomy samples via laser capture microdissection (GSE41571)36. F-H) Previous work on atherosclerotic aortas used scRNA-seq to detect leukocyte subsets along with their signature genes. Heatmaps show the relative expression of the identified signature genes in each of the 4 fluorescent subsets found here, with each gene normalized to its own minimum (blue) and maximum (red) normalized count, averaged by group. Genes within each of the predefined subsets were allowed to cluster freely. F) Cochain, et al., identified three aortic macrophage subsets in LDLr−/− mice- inflammatory, resident-like (Res-like), and Trem2hi- with 4 signature genes each11. G) Lin, et al., reported 8 macrophage subsets amongst all leukocytes in LDLr−/− mice. 4–5 signature genes were provided for each cluster. H) Kim, et al., described 11 cluster of CD11b+ cells derived from CX3CR1+ monocytes13. 20 overexpressed genes were provided for each cluster.
