Figure 6. New pS22-Lamin A/C-binding sites emerged in progeria are associated with up-regulation of genes relevant to progeria phenotypes.
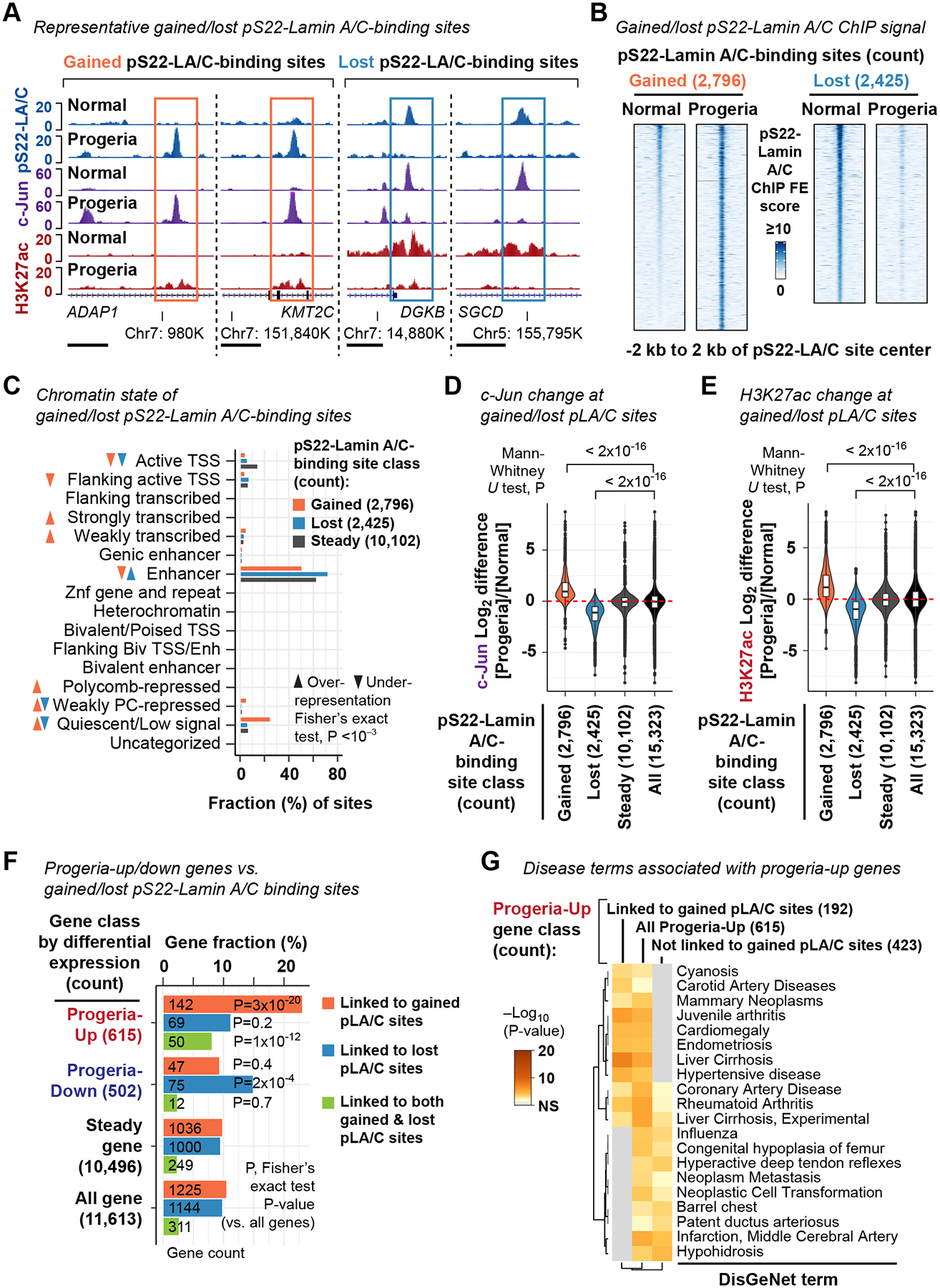
(A) pS22-Lamin A/C ChIP-seq profiles (derived from 2 biological replicates) at representative gained and lost pS22-Lamin A/C-binding sites. Normal, GM07492 fibroblast. Progeria, AG11498 fibroblast. Horizontal bar, 2 kb. See also Fig. S6.
(B) pS22-Lamin A/C ChIP-seq FE scores at gained and lost pS22-Lamin A/C-binding sites in the normal GM07492 and progeria AG11498 fibroblasts.
(C) Chromatin states of gained and lost pS22-Lamin A/C-binding sites. Fisher’s exact test assesses association between being called as gained or lost and being affiliated with each state.
(D) Log2 difference of c-Jun ChIP-seq FE scores between normal (GM07492) and progeria (AG11498) fibroblasts (derived from 2 biological replicates). Red dashed line, log2 difference equals 0 for reference.
(E) Same as (D), but H3K27ac ChIP-seq scores are analyzed.
(F) Fraction of progeria-up and progeria-down genes with gained and/or lost pS22-Lamin A/C-binding sites (within gene body or 100 kb upstream). P, Fisher’s exact test P value for association between being differentially expressed and being affiliated with gained/lost pS22-Lamin A/C-binding sites.
(G) DisGeNet-curated disease terms over-represented among progeria-up genes linked to gained pS22-Lamin A/C-binding sites, or those not linked to gained pS22-Lamin A/C-binding sites, or all progeria-up genes.
