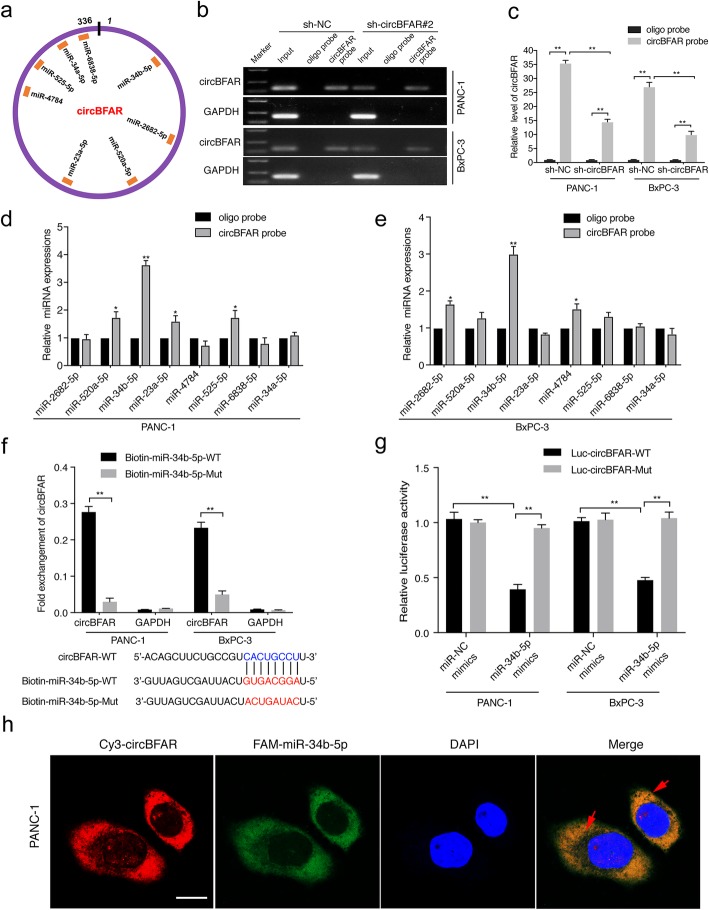
Fig. 4.
CircBFAR serves as a sponge for miR-34b-5p in PDAC cells. a Schematic illustration showing potential target miRNAs of circBFAR as predicted by miRanda, starbase, and RNA hybrid. b, c The specificity and efficiency of the circBFAR probe was validated using gel electrophoresis and qRT-PCR in PANC-1 and BxPC-3 cells. d, e qRT-PCR analysis of the expression of eight potential target miRNAs in PANC-1 and BxPC-3 cells. MiR-34b-5p was stably pulled down by circBFAR in PANC-1 and BxPC-3 cells. f Biotinylated miRNA pull-down (WT or mut) and qRT-PCR assays showing the expression levels of circBFAR after co-transfection of circBFAR and miR-34b-5p mimics in PANC-1 and BxPC-3 cells. GAPDH was used as the negative control. g The luciferase activities of the circBFAR luciferase reporter vector (WT or mut) measured after transfection with miR-34b-5p mimics or mimic NC into PANC-1 and BxPC-3 cells. h The co-localization of circBFAR and miR-34b-5p in PANC-1 cells was detected using a FISH assay. CircBFAR probes were labeled with Cy3. miR-34b-5p probes were labeled with FAM. Nuclei were stained with DAPI. The images were photographed at 1000X magnification. Scale bar = 10 μm. Statistical significance was assessed using two-tailed t-tests for two group comparison, and one-way ANOVA followed by Dunnett’s tests for multiple comparison. The error bars represent the standard deviations of three independent experiments. *P < 0.05, **P < 0.01