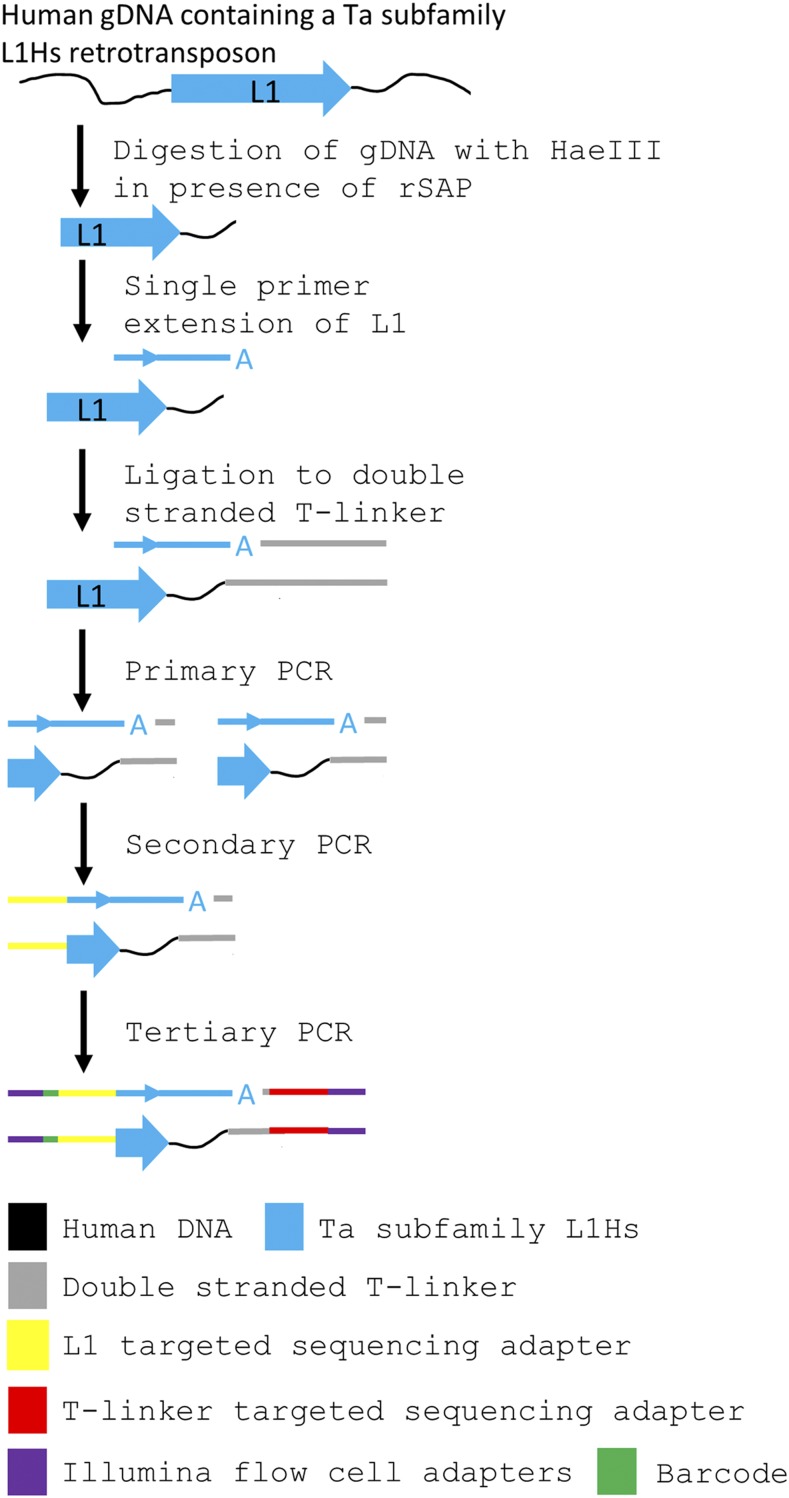
Figure 2.
Schematic of the construction of Ta subfamily enriched L1Hs sequencing libraries. gDNA isolated from NeuN+ nuclei was enzymatically digested with HaeIII in the presence of shrimp alkaline phosphatase (rSAP) to fragment the genome and remove 5′ phosphates from cleavage products. A single primer extension using the Ta subfamily specific L1HsACA primer extends the 3′ end of the L1 sequence into the downstream gDNA. The 3′ ‘A’ overhang from the single primer extension is ligated to a custom T-linker, and primary PCR amplifies the construct using L1HsACA and T-linker specific primers. Hemi-nested secondary PCR using the L1Hs specific L1HsG primer and T-linker primer reduces the length of the L1 sequence carried forward and adds a sequencing adapter to the L1 end. Tertiary PCR uses primers complementary to the 5′ end of library amplicons to add a barcode to the L1 end and Illumina flow cell adapters to both ends of the amplicons.