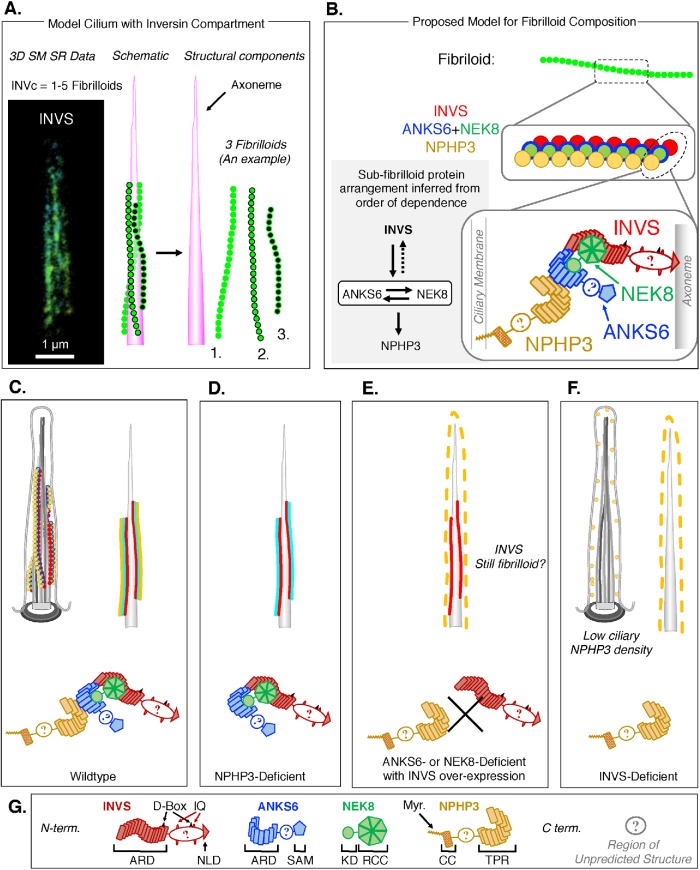
FIGURE 6:
A new model for the structure and composition of the INVc. (A) 3D SM SR reconstructions of INVS (left) suggest that the INVc is composed of fibrilloid substructures. Based on these data, a schematic of a cilium with an INVc, which is composed in this example of three fibrilloids (green) that are aligned roughly parallel along the axoneme (magenta), is shown. (B–G) Illustrations of the structure and composition of the INVc at various levels of detail and in different genetic contexts (C–F). Each INVc protein is shown in following colors: INVS (red), ANKS6 (blue), NEK8 (green), and NPHP3 (yellow). In the highest-resolution illustrations, each INVc protein is illustrated with some structural details pertaining to predicted protein domains within each molecule (defined in G). (B) A possible model for fibrilloid composition based on the order of dependence for the assembly of the four INVc proteins in the compartment. The 1:1:1:1 stoichiometry for all INVc proteins shown here is symbolic, and the relative positions of the circular molecules in the middle-resolution illustration are intentionally ambiguous as to whether each fibrilloid is a heteropolymer of all or some INVc proteins or a homopolymer, consisting for example of INVS only, on which ANKS6-NEK8 and NPHP3 then assemble (see Supplemental Figure S59 for candidate polymer models). In the highest-resolution illustrations (bottom), NPHP3 associates directly with the ciliary membrane via its N-terminal myristoylation, and ANKS6–NEK8 complexes link NPHP3 to INVS. (C) Illustrations of a cilium in a WT cell in which the INVc is a fibrilloid structure composed of INVS, ANKS6, NEK8, and NPHP3. Three fibrilloids are shown in the more detailed illustration (top left). Two fibrilloids are shown in the simplified illustration (top right), where ANKS6 and NEK8 are together represented in cyan. INVS is the core organizer and is illustrated here as being more directly associated with the axoneme. (D) A cilium in an NPHP3-deficient cell. The INVc is still present and structurally unaffected by loss of NPHP3. (E) A cilium in an ANKS6- or NEK8-deficient cell in which GFP-INVS expression has rescued compartment formation. The INVc contains INVS, which we expect still forms fibrilloids. Ciliary NPHP3 density is reduced, and NPHP3 is not sequestered in the INVc, but rather relocalized throughout the ciliary volume. The failure of INVS to sequester NPHP3 is due to the loss of the ANKS6–NEK8 complex that is predicted to link these two proteins physically, based on our measurements of NPHP3 localization in ANKS6-KO or NEK8-KO cells stably expressing GFP-INVS. (F) A cilium in an INVS knockout cell. No INVc is present. NPHP3 is reduced and found throughout the cilium, albeit at a low density, similar to that observed for ANKS6- or NEK8-deficient cells. (G) Schematic of the protein domains and motifs present in the four INVc proteins as cartooned in panels B–F. All four proteins are oriented left to right from N- to C-terminus. The INVS protein contains an ankyrin repeat domain (ARD) consisting of 16 ankyrin repeats, two D-boxes (known to bind APC; Morgan et al., 2002), a bipartite NLS (Otto et al., 2003), two IQ domains (known to bind calmodulin; Yasuhiko et al., 2001), and a C-terminal ninein-like domain (NLD) that is important for centriolar targeting of INVS (Shiba et al., 2009) ANKS6 contains an N-terminal ARD with 11 ankyrin repeats, followed by a serine-rich patch and a C-terminal SAM domain. The ANKS6 ARD likely mediates an interaction with NEK8 through the N-terminal NEK8 kinase domain (KD; Czarnecki et al., 2015). NEK8 contains a C-terminal RCC (regulator of chromatin condensation) domain, which is predicted to fold into a propeller structure and is important for mediating an interaction of NEK8 and INVS (Zalli et al., 2012; Czarnecki et al., 2015). The N-terminus of NPHP3 is myristoylated (Myr.), which could insert in the ciliary membrane (Wright et al., 2011; Nakata et al., 2012; as illustrated in panel B). NPHP3 also contains a coiled-coil domain (amino acids 83–207), which may be important for targeting NPHP3 to the basal body (Nakata et al., 2012). The C-terminus of the NPHP3 contains at least 11 tetratricopeptide repeats (TPR). The first 200 amino acids of NPHP3 localize to the cilium (Wright et al., 2011) but fail to be sequestered in the INVc (Supplemental Figure S37), suggesting that the interaction between NPHP3 and ANKS6 or NEK8 is either through the TPR repeats or through some other domain present in the C-terminus of the protein (amino acids 201–1330).