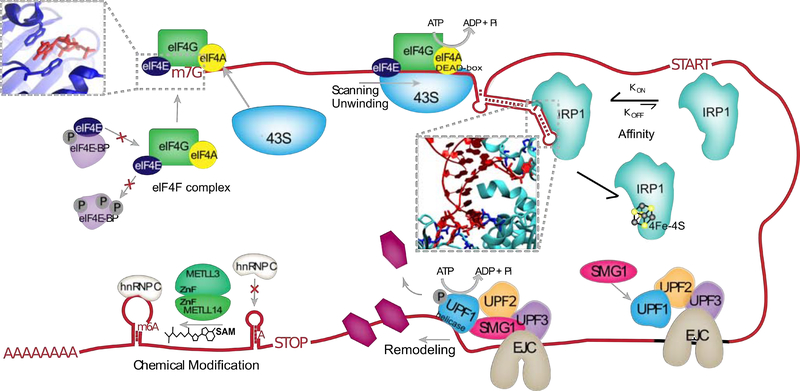
Figure 5.
Examples of mechanisms controlling RBP binding, interactions with RNA, and their regulation. eIF4E (dark blue) interacts with 7-methyl-guanosine cap (m7G), in part through stacking interactions (inset) and binds to RNA as part of the eIF4F which includes RBPs eIf4G and eIF4A. eIF4E association with eIF4F is prevented by sequestration to hypo-phosphorylated eIF4E-BP. The 43S ribosomal subunit is recruited to the eIF4F complex, and processively scans the 5’UTR, aided by ATP-driven helicase activity of the eIF4A DEAD-box domain. The RBP IRP1 specifically binds hairpin elements in the 5’UTR with high affinity through specific residues (inset, dark blue) that hydrogen bond with the bulge and apical loop of the RNA. RNA binding by IRP1 is prevented by 4Fe-4S ligand binding to IRP1. UPF1 is recruited to the exon junction complex (EJC), where its helicase activity is activated by interactions with SMG1 and UPF2. Driven by ATP, UPF1 removes both RNA structures and other bound RBPs in the 5’−3’ direction. A zinc-finger (ZnF) containing METTL3-METTL14 complex deposits methyl groups donated by S-adenosyl methionine (SAM) on targeted adenosines (m6A). M6A modifications reduce base-pairing in RNA, such that some locations become available for hnRNPC binding.