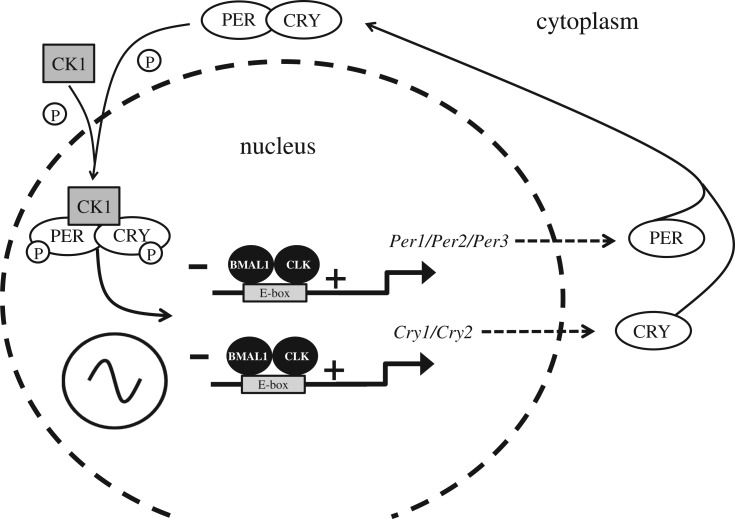
Figure 2.
The mammalian molecular clock. The driving force of the mammalian molecular clockwork is transcriptional/translational feedback loop (TTFL) the transcriptional drive is provided by two proteins named ‘Circadian Locomotor Output Cycles Kaput’, or less torturously CLOCK (CLK), which links with ‘Brain muscle arnt-like 1’ or BMAL1. The CLK–BMAL1 complex binds to E-box promoters driving transcription of five core clock genes, three Period genes (Per) giving rise to the proteins PER1, PER2 and PER3, and two Cryptochrome genes (Cry) which encode the CRY1 and CRY2 proteins. The PER proteins combine with the kinase CK1 (Casein kinase 1) and are phosphorylated. The PER–CK1 complex then binds to the CRYs to form a CRY–PER–CK1 complex. Within the complex of CRY–PER–CK1, CRY and PER are phosphorylated by other kinases which then allows the CRY–PER–CK1 complex to move into the nucleus and inhibit CLK–BMAL1 transcription of the Per and Cry genes forming the negative limb of the TTFL. The CRY–PER–CK1 protein complex levels rise throughout the day, peak at dusk, are then degraded and decline to their lowest level the following dawn. The net result is a TTFL, whereby the Per and Cry genes and their protein products interact and feedback to inhibit their own transcription, generating a 24 h cycle of protein production and degradation. Note, multiple other genes and their proteins, generate additional feedback loops to provide further stability to the circadian oscillation [8]. Significantly, polymorphisms in several of these clock genes have been associated with human ‘morning types’ (larks) and ‘evening types’ (owls) [9].