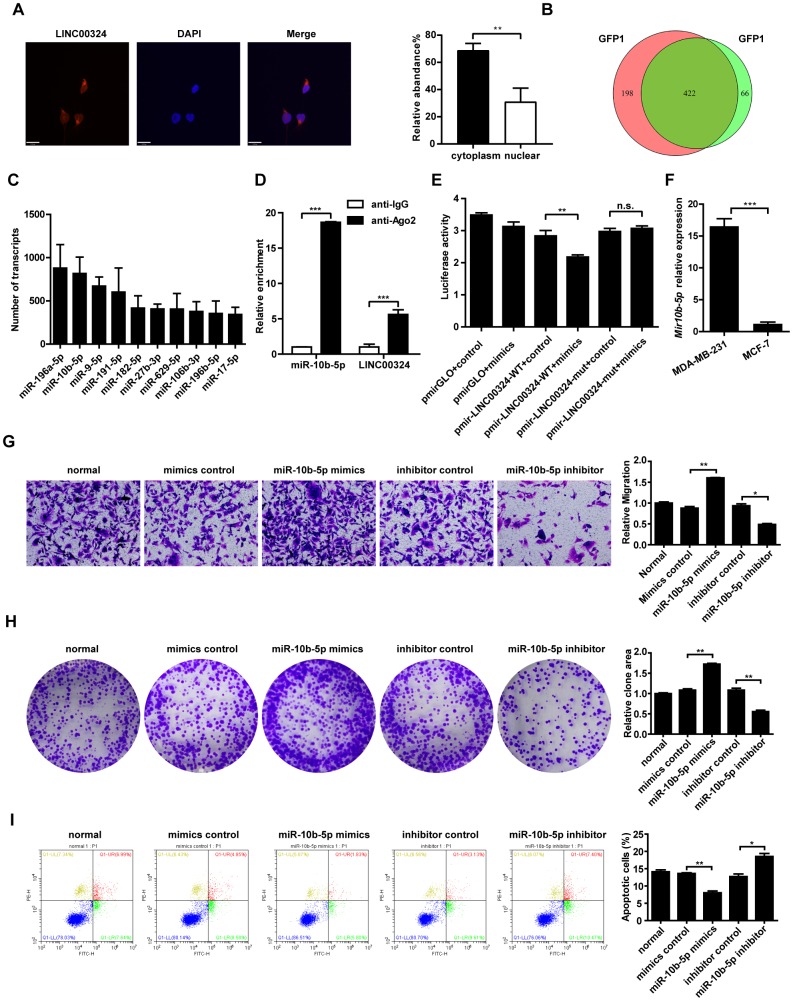
Figure 5.
Candidate miRNAs scanning, verification, and function correlation with LINC00324. (A) FISH assay was performed to detect the subcellular distribution of LINC00324 in MDA-MB-231 cells. The LINC00324 probe mix and control RNA probe mix were labeled with Cy3. DAPI was used to counterstain the nuclei. The high resolution images were captured with a laser scanning confocal microscope. (B) Wayne diagram indicates the overlap microRNA transcrips of two independent groups. (C) Top ten potential LINC00324 bound reads from MS2-RIP-seq. (D) RIP assays showing the association of LINC00324 with miR-10b-5p in MDA-MB-231 cells. (E) MDA-MB-231 cells were co-transfected with LINC00324- 3′ UTR wild type or LINC00324- 3′ UTR mutation reporter plasmids, together with miR-10b-5p mimic or mimic negative control and then subjected to the luciferase assay. (F) Relative expression of miR-10b-5p in MDA-MB-231 and MCF-7 cells. (G) Migration ability of MDA-MB-231 cells determined by Transwell assay. (H) Colony formation assays performed with MDA-MB-231 cells transfected with miR-10b-5p mimics, miR-10b-5p inhibitor, or negative control. (I) Flow cytometry analysis of the percentage of apoptotic MDA-MB-231 cells with miR-10b-5p mimics or miR-10b-5p inhibitor. All data are shown as means ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001. Data are from three independent experiments (D–F), or are representative of three independent experiments with similar results (A, G, H, I).