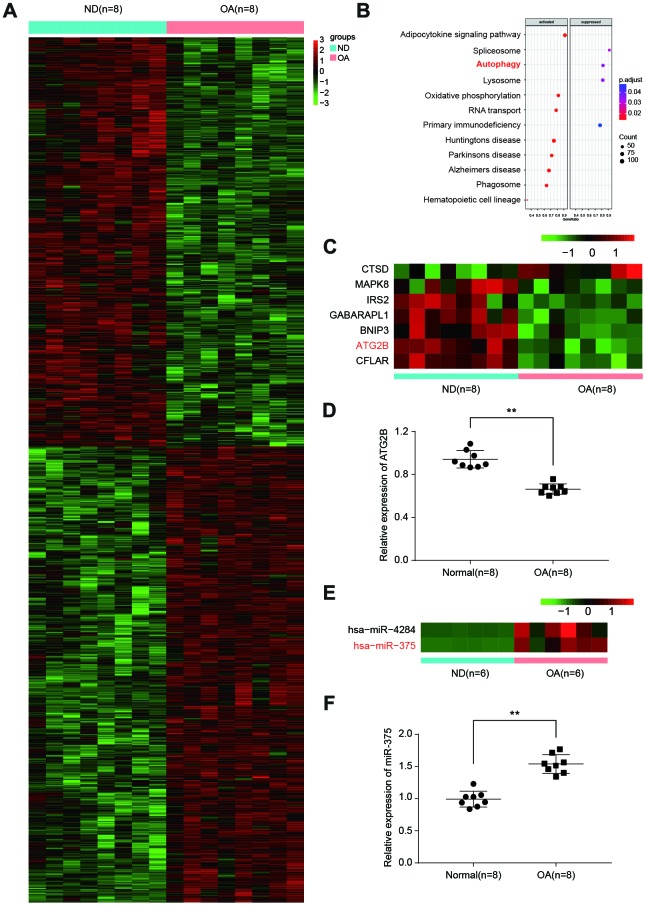
Figure 1.
Differential expressed mRNAs and miRNAs related to autophagy in OA. (A) Heatmap of top differentially expressed mRNAs from 8 matched osteoarthritic and normal samples in osteoarthritis (OA) patients. (B) Significant function and pathways (P<0.05) based on KEGG database were analyzed using GSEA tool from upregulated and downregulated genes. Activated pathways were indicated in the left box, with suppressed pathways showing in the right box. (C) Heatmap depicting statistically significant (P<0.05) differentially expressed mRNAs related to autophagy. (D) Relative expression of ATG2B in osteoarthritic and normal samples was analyzed by qRT-PCR. (E) Heatmap depicting differentially expressed miRNAs related to autophagy from 6 matched osteoarthritic and normal samples. (F) Relative expression of miR-375 was analyzed by qRT-PCR.