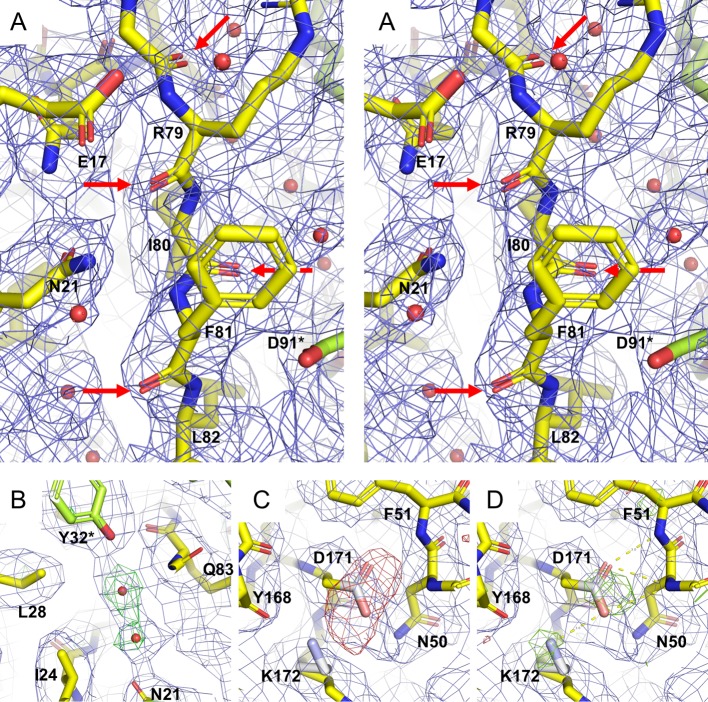
Fig 2. Representative densities during quasi-crystallographic refinement against the cryo-EM Coulomb potential map.
(A) Stereo view demonstrating cryo-EM Coulomb potential map density (blue cage) for carbonyl oxygens (red arrows) in accordance with a nominal resolution of 2.7 Å. The final refined model (PDB code 6SHT) is shown as yellow sticks, with octahedrally symmetry related residues colored lime. (B) Positive difference electron density (Fmap-Fcalc), green, contoured at 2.5σ) after first round of quasi-crystallographic refinement of manually fitted apoferritin monomer superimposed with the final model (yellow / lime sticks). As the initial refinement was based on protein residues alone, the difference density clearly demonstrates the presence of two solvent molecules, also present in the cryo-EM map. These water molecules are also present in the crystal structure (PDB code 3WNW). (C) The initial refinement also exhibited negative difference density (red cage, contoured at -3.5σ) for all carboxylate groups (here Asp171). These atoms (Asp: Cγ, Oδ1 and Oδ2: Glu: Cδ, Oε1 and Oε1) were given an occupancy of zero so that they no longer contribute to structure factor calculations (i.e. set dummy, white / pink atoms). For a number of carboxylate groups including Asp171, the next round of refinement revealed positive difference density (D, green cage contoured at 3.5σ), demonstrating an influence of these atoms on the cryo-EM Coulomb potential. This round of refinement also revealed density for a second conformation of the Lys172 side chain (white sticks).