Fig. 1. The FOXP3 locus is precisely targeted using the CRISPR system in primary HSPCs and T cells.
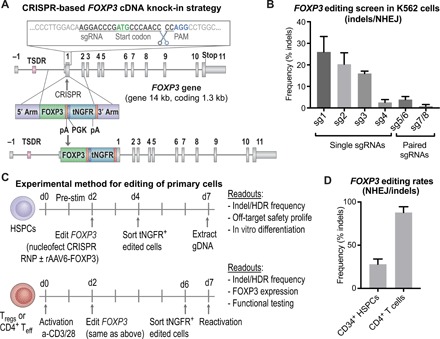
(A) Schematic representation of CRISPR-based editing of the FOXP3 gene showing the CRISPR cut site in first coding exon, E1 (exons depicted by gray boxes separated by lines representing introns; the first coding exon, E1, is preceded by the noncoding exon E-1 and the enhancer with TSDR). A zoomed-in view of the sgRNA binding site relative to the start codon, PAM site, and cleavage site is shown. Homology donor depicted below with arms of homology, codon divergent FOXP3 cDNA, BGH polyadenylation (pA) signal included to terminate the FOXP3 transcript, truncated NGFR (tNGFR) marker gene under the Phosphoglycerate Kinase (PGK) promoter to drive marker expression independent of FOXP3 expression, and a second pA. (B) Screening of sgRNAs targeting the first coding exon of the FOXP3 gene. Plasmids encoding WT Cas9 or nickase variant of Cas9 (paired sgRNAs) and FOXP3 sgRNAs nucleofected into K562 cell lines. CRISPR efficiency measured by TIDE analysis to detect insertion deletion (indel) mutations created by nonhomologous end joining (NHEJ)–mediated DNA repair. (C) Experimental method for editing of HSPCs and T cells with functional readouts listed. (D) CRISPR cutting efficiency in CD34+ HSPCs and CD4+ T cells quantified by TIDE analysis for the detection of indel mutations created by the NHEJ repair pathway.
