Fig. 6. FOXP3 edited HSPCs undergo multilineage hematopoietic differentiation and engraftment in vitro and in vivo.
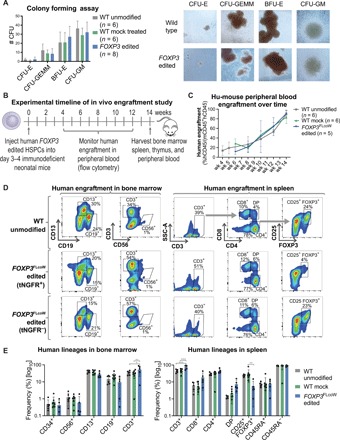
(A) Differentiation potential of edited HSPCs tested by the in vitro CFU assay. Four resulting hematopoietic progenitor colony types: CFU-E (mature erythroid progenitors), CFU-GEMM (granulocyte, erythrocyte, macrophage, and megakaryocyte), BFU-E (primitive erythroid progenitors), and CFU-GM (granulocyte and macrophage progenitors). Representative images of colonies from the CFU assay, showing similar morphology (×10 magnification). (B) Experimental timeline of hu-mouse study using NSG-SGM3 mice. (C) Human engraftment kinetics in the peripheral blood of hu-mice at corresponding weeks after injection. Engraftment was measured by flow cytometry for hCD45 marker on human cells, and frequency was quantified relative to the total of human (hCD45+) and mouse (mCD45+) cells (mean ± SD). (D) Representative flow cytometry plots of engrafted human hematopoietic subsets in the bone marrow (left) and spleen (right) of hu-mice at 14 weeks after injection. Populations gated out of human cells (hCD45+). FOXP3 edited samples were divided into tNGFR+ and tNGFR− gates for comparability. (E) Quantification of human hematopoietic lineages by flow cytometry with each symbol representing a single mouse (mean ± SD). In spleen, the CD8+, CD4+, and CD4+CD8+ double-positive (DP) populations were gated out of CD3+ T cells. The CD25+FOXP3+, naïve CD45RA+, and memory CD45RA− populations were gated out of CD4+ single-positive T cell subset (*P < 0.5, **P < 0.01, ***P < 0.001).
