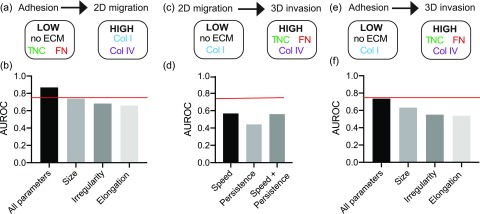
FIG. 5.
Adhesion classifies ECM-driven 2D migration and 3D invasion. (a) Cell adhesion to predict binary classification of 2D cell migration speed of MDA-MB-231 cells on plastic, Collagen I, Fibronectin, Tenascin C, or Collagen IV. (b) AUROC scores of binary AdaBoost classifier models (a) using all 11 cell shape parameters, cell size parameters (area/cell, perimeter, mean radius, min feret diameter, max feret diameter), cell irregularity parameters (solidity, extent, form factor), and cell elongation parameters (eccentricity, aspect ratio, compactness). (c) 2D cell migration to predict binary classification of mean fold change of spheroid area of 231-GFP cells embedded in media, Collagen I, Fibronectin, Tenascin C, or Collagen IV. (d) AUROC scores of binary classifier models (c) using 2D cell migration (cell migration speed and persistence alone or together). (e) Cell adhesion to predict binary classification of mean fold change of spheroid area of 231-GFP cells embedded in media, Collagen I, Fibronectin, Tenascin C, or Collagen IV. (f) AUROC scores of binary classifier models (e) using all 11 cell shape parameters, cell size parameters, cell irregularity parameters, and cell elongation parameters.