Figure 7.
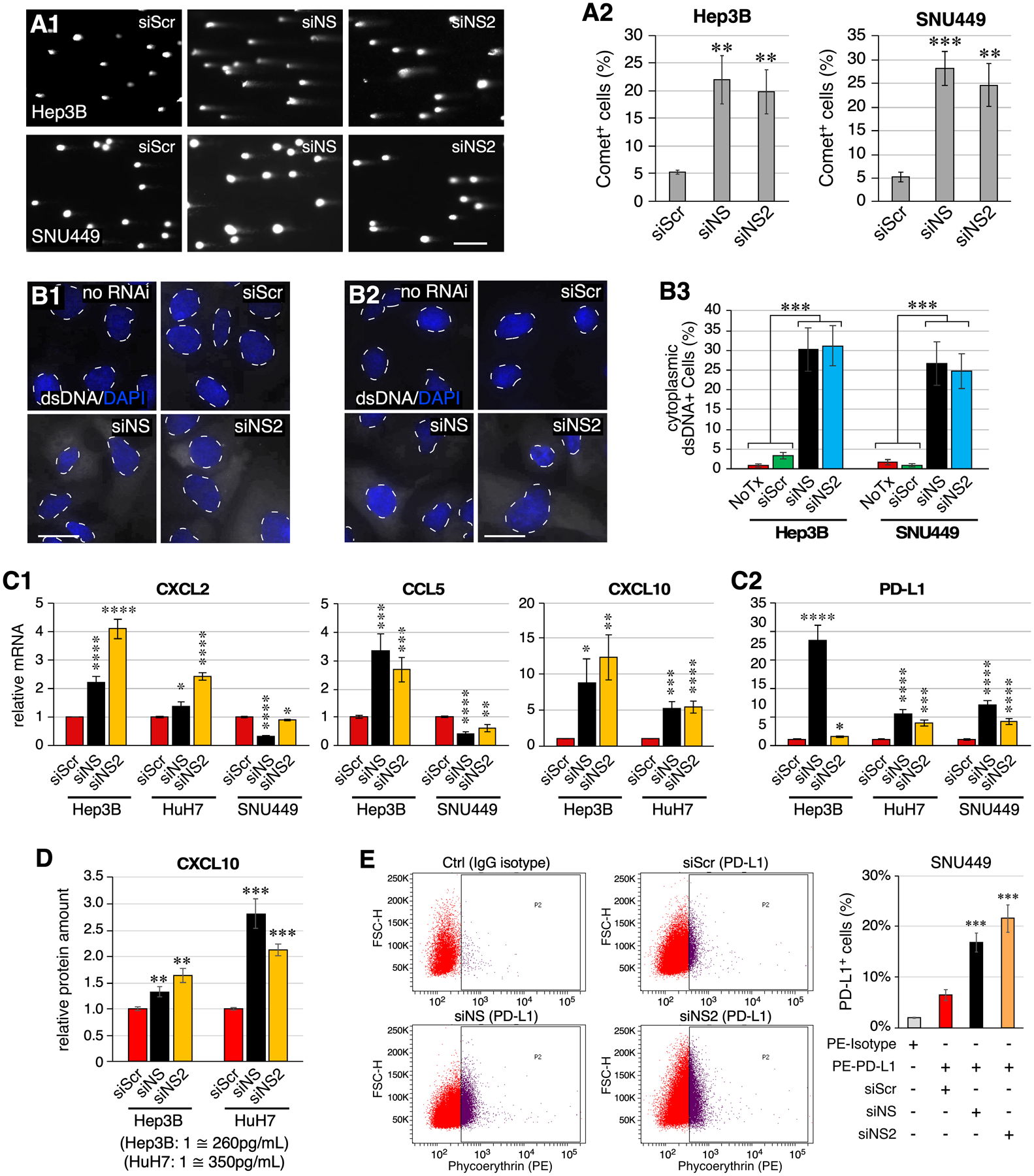
NS depletion leads to increased cytosolic double-stranded DNAs and triggers innate immune response in HCC cells. (A) Images and analyses of DNA tail moments of RNAi-treated Hep3B and SNU449 cells by Comet assay. (B) Images of dsDNA staining in no-RNAi and RNAi-treated Hep3B cells (B1) and SNU449 cells (B2). Signals of dsDNA and DAPI are shown in white and blue colors, respectively. Nuclear-cytoplasmic borders are demarcated by dashed lines. Statistical analyses are shown in (B3). (C) Expression levels of CXCL2, CCL5, CXCL10, and PD-L1 in RNAi-treated Hep3B, HuH7, and/or SNU449 cells by qRT-PCR. Levels were referenced to Rplp0 in each sample and compared to their respective siScr controls. (D) ELISA analysis of CXCL10 in the medium collected from RNAi-treated Hep3B and HuH7 cell cultures. Data consist of four biological repeats with two technical replicates. (E) FACS analysis of PD-L1+ cell percentage of RNAi-treated SNU449 cells labeled with PE-conjugated anti-PD-L1 antibody. Untreated SNU449 cells labeled with PE-conjugated IgG isotype antibody were used as the gating control. Scale bars, 200μm in (A) and 20μm in (B). The NSKD responses of CCL5 in HuH7 cells, CXCL10 in SNU449 cells, and STING in Hep3B cells were not included because of their low baseline expression levels in those cell lines, limiting the reliability of qRT-PCR results.
