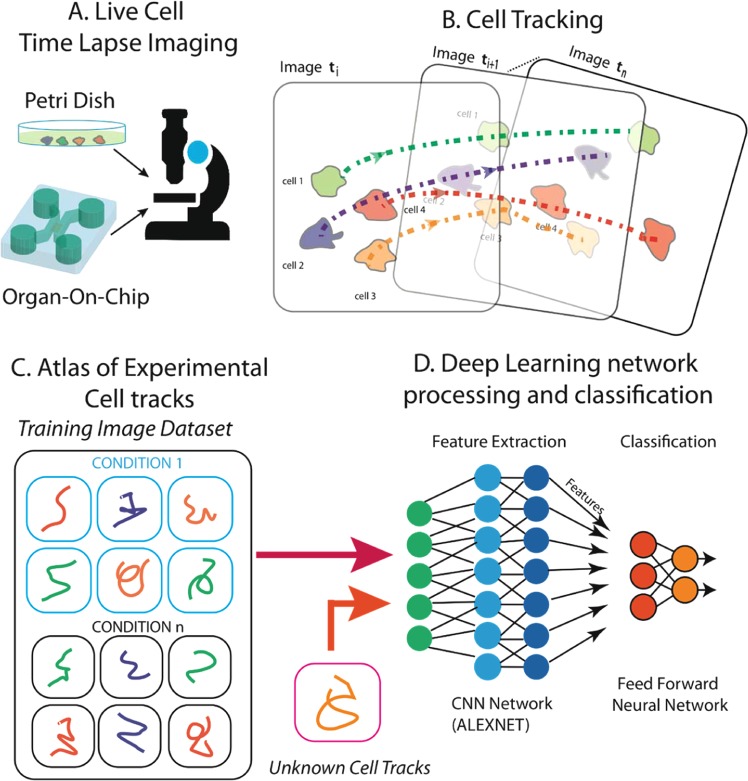
Figure 1.
A schematic representation of the proposed method. (A) Time-lapse microscopy is used to acquire the video sequence of cells moving in a Petri dish or in an OOC platform. (B) Cells are localized and tracked through the video sequence. (C) For each cell (or cluster of cells) of interest, an atlas of trajectories is collected for the different biological conditions under consideration. (D) Through a pre-trained Deep Learning architecture, i.e., AlexNET, the tool provides a feature representation of each ATLAS in order to perform experiment classification through a predefined taxonomy (e.g., drug vs no-drug).