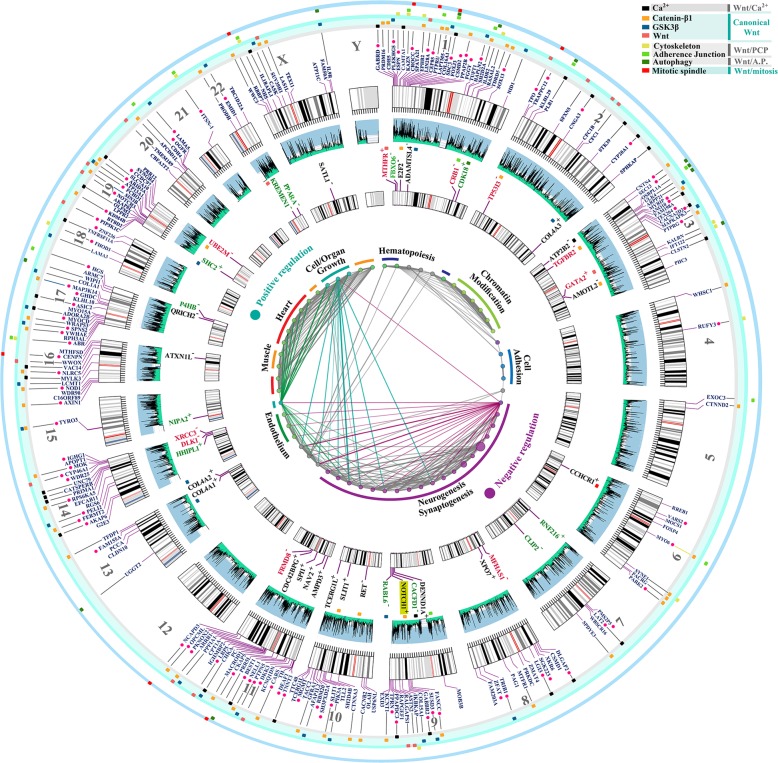
Fig. 4. Spatial and functional clustering of homology to miR-4673.
Genomic distribution of miR-4673 interactome shows distinct gene clusters localised to H3+ isochores (middle circular diagram). The miRNA homology regions (miRHR) were scattered in coding sequences (inner karyogram genes in black), 5′e-UTRs (inner karyogram genes in red), 3′e-UTRs (inner karyogram genes in green) and i-UTRs of human genes (outer karyogram). Sense and anti-sense directionality of (miRHR) in the inner karyogram is designated as + and −. In the outer karyogram red circles indicate the genes with miRHR in antisense direction to miR-4673. Genes with miRHR signature are involved in regulating development of cardiovascular and nervous systems (inner diagram). These genes act as calibrators (tissue-specific modulators) of Wnt signalling by fine-tuning canonical and non-canonical modules of the cascade (see Supplementary Discussion 1 for detailed elaboration). The outer colour-coded layer designates the interactome genes to canonical Wnt and non-canonical Wnt/Ca2+, Wnt/PCP (planar cell polarity), Wnt/Autophagy (A.P.) and Wnt/Mitosis cascades.