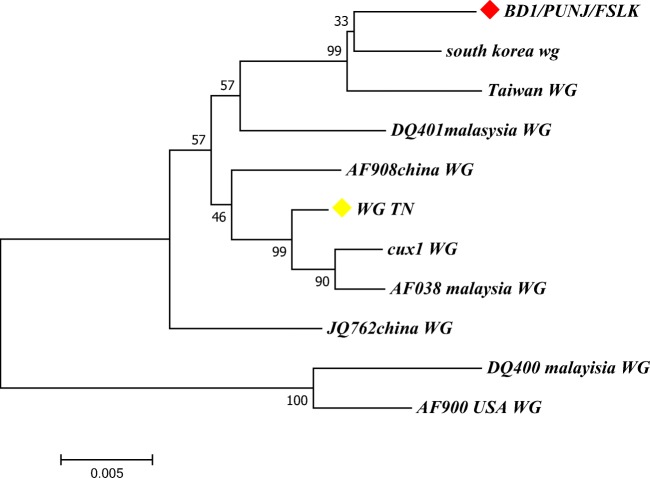
Fig. 6.
Molecular phylogenetic analysis of CIAV genome. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura-Nei model. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analysed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. BD1/PUNJ/FSLK grouped with South Korea and Taiwan with bootstrap value of 99