FIGURE 2.
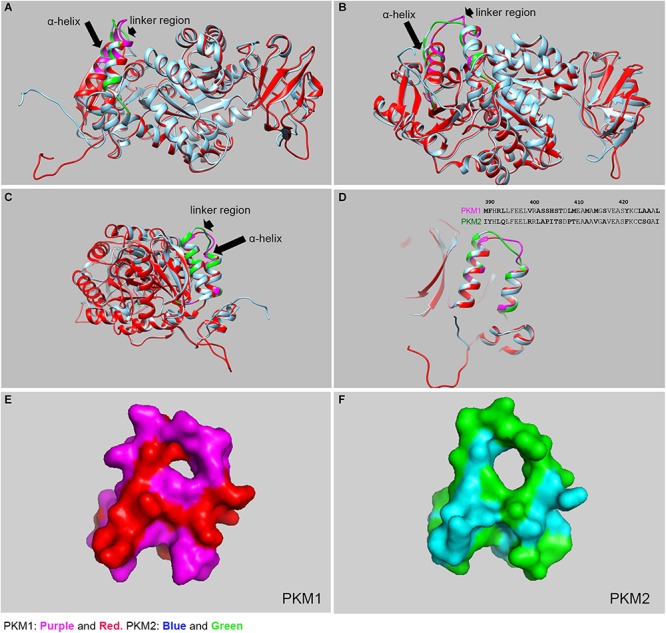
Structural comparison between mouse PKM1 and PKM2. The crystal structure of human PKM1 and PKM2 has been solved (PDB ID: PKM2, 4JPG, PKM1, 3SRF). We carried out the molecular modeling of mouse PKM1 and PKM2 using the Raptor X template-based protein structure modeling web server and aligned, with Clustal Omega, and the resulting structures were generated using the UCSF Chimera program and PyMOL (PyMOL Molecular Graphics System, Version 4.5 Schrödinger, LLC). The structural alignment of PKM1 and PKM2 at the different views (A–C). The purple color represents PKM1 divergent region/residue and the red represents PKM1 conserved region/residue. The green color represents the PKM2 divergent region/residues and the blue represents PKM2 conserved region/residue. The majority of residues in PKM1 are identical to PKM2, except a region where diverge can be noticed in the linker region (see arrowhead) and 2 alpha helices (see arrow). A significant divergence was shown to be localized to residues 389–428 which consist of 2 alpha helices and joining linker region (D). inset: Divergent protein sequence between PKM1 and PKM2 (389–428). The surface plots of PKM1 (E) and PKM2 (F), respectively as seen in panel (D).
