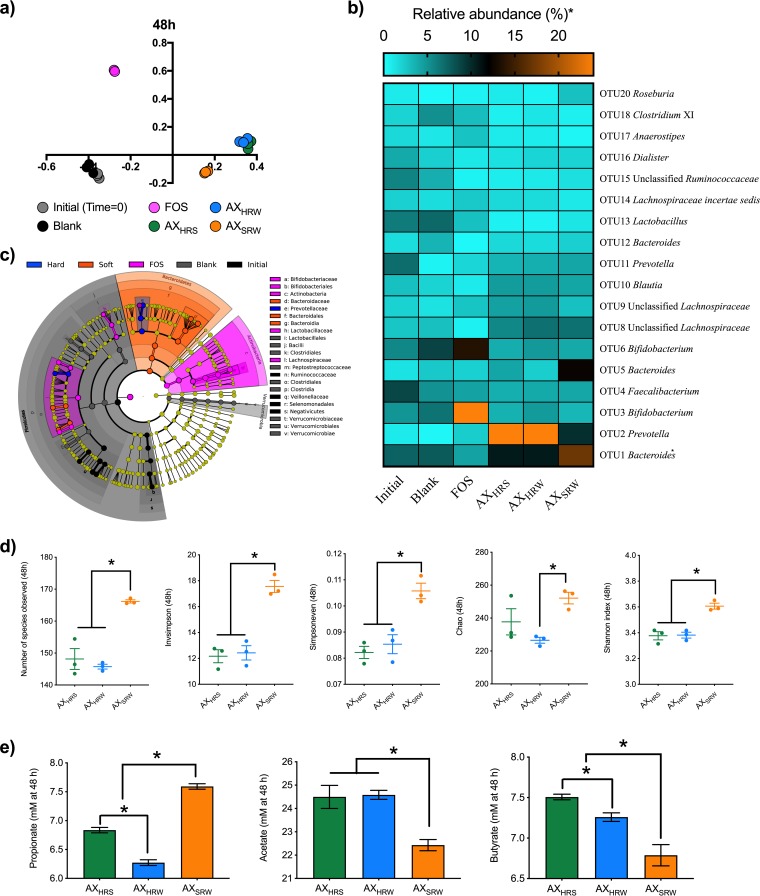
FIG 2.
Microbial community analyses after in vitro fermentation for 48 h, as determined by 16S rRNA gene amplicon sequencing. (a) Bray-Curtis dissimilarity of fecal microbial communities based on the relative abundances of OTUs at 97% similarity level (principal-component analysis [PCA] plots for the 12- and 24-h time points are given in Fig. S2). Dissimilarity was also calculated using ThetaYC; the result was not substantially different from that visualized by Bray-Curtis dissimilarity. (b) Relative abundances (percentage of sequences) based on the top 50 OTUs in each sample. The top 50 OTUs accounted for more than 90% of the total sequences of all AX treatment groups at all time points (relative abundances at the 12- and 24-h time points are given in Fig. S4, and relative abundances displayed in a bar graph are provided in Fig. S5). (c) Cladogram (obtained as a result of linear discriminant analysis) depicting taxa that are overrepresented in the AX samples obtained from hard and soft wheat classes compared with abundances in the initial inoculum and substrate-free blank incubations. (d) Changes in α-diversity of the fecal microbiota communities, as measured by number of species observed and inverse Simpson, Simpson, Chao, and Shannon’s index calculators (α-diversity of the fecal microbiota communities at the 12- and 24-h time points are given in Fig. S3). (e) Short-chain fatty acid (SCFA; namely, propionate, acetate, and butyrate) production by fecal microbiota at the end of the fermentation (the amounts of SCFAs produced after in vitro fermentation for 12 and 24 h are given in Fig. S6a; the proportions of all SCFAs produced after in vitro fermentation for 12, 24, and 48 h are given in Fig. S6b). Fructooligosaccharide (FOS) was used as a fast-fermenting, butyrate-producing positive control. The blank did not contain any substrate. Statistical analyses were done using two-tailed Student’s t test. Error bars represent the standard errors of three separate replicates.