Fig. 2.
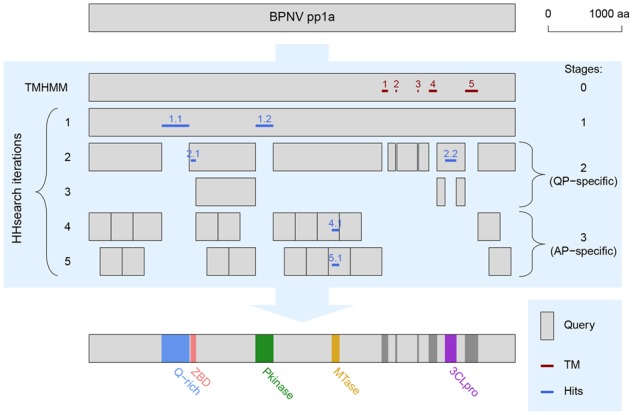
LAMPA workflow and its application to RNA virus polyprotein. Presented is outline of the LAMPA approach (blue background) applied to polyprotein 1a (pp1a) of BPNV. Gray bars, regions of BPNV pp1a that served as TMHMM or HHsearch queries. Iterations of the procedure and programs used are depicted on the left; stages are indicated on the right. Clusters of TM helices are depicted in dark red, clusters of hits—in dark blue. Hit double digits refer to iteration and hit position on polyprotein from left to right, respectively, except for hits at Stage #0 which are labelled with the position only. Hits and annotations obtained on Stage #1 represent output of conventional HHsearch. Q-rich, region rich in glutamine residue; ZBD, zinc-binding domain; Pkinase, protein kinase; MTase, methyltransferase; 3CLpro, 3C-like protease. For other details see text. (Color version of this figure is available at Bioinformatics online.)
