Fig. 5.
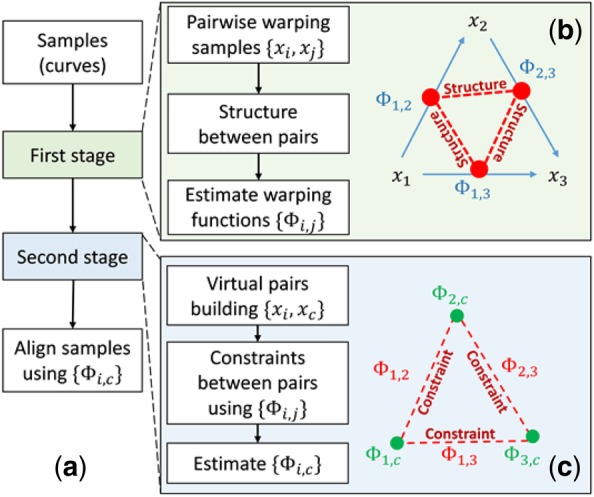
Flowchart of ncGTW algorithm. (a) With two-stage alignment strategy, all input samples (curves) are aligned simultaneously to a virtual reference. (b) Stage 1 of ncGTW with three illustrative samples. First, ncGTW builds a pairwise warping flow map (blue arrows). Then ncGTW incorporates structural information as the constraint and applies to all pairs (pair as red dot and constraint as a red dashed line). Lastly, ncGTW estimates all pairwise warping functions (Φ_(i,j)) jointly with e.g. smoothness constraint on neighboring sample pairs. (c) Stage 2 of ncGTW with three illustrative samples. ncGTW aligns every sample to a common virtual reference x_c, where the warping functions {Φ_(i,j)} obtained in Stage 1 provide warping correspondences and final warping functions {Φ_(i,c)} are calculated by solving the maximum flow problem. (Color version of this figure is available at Bioinformatics online.)
