Figure 1.
GPT1 Reporter Fusions Dually Localize to Plastids and the ER.
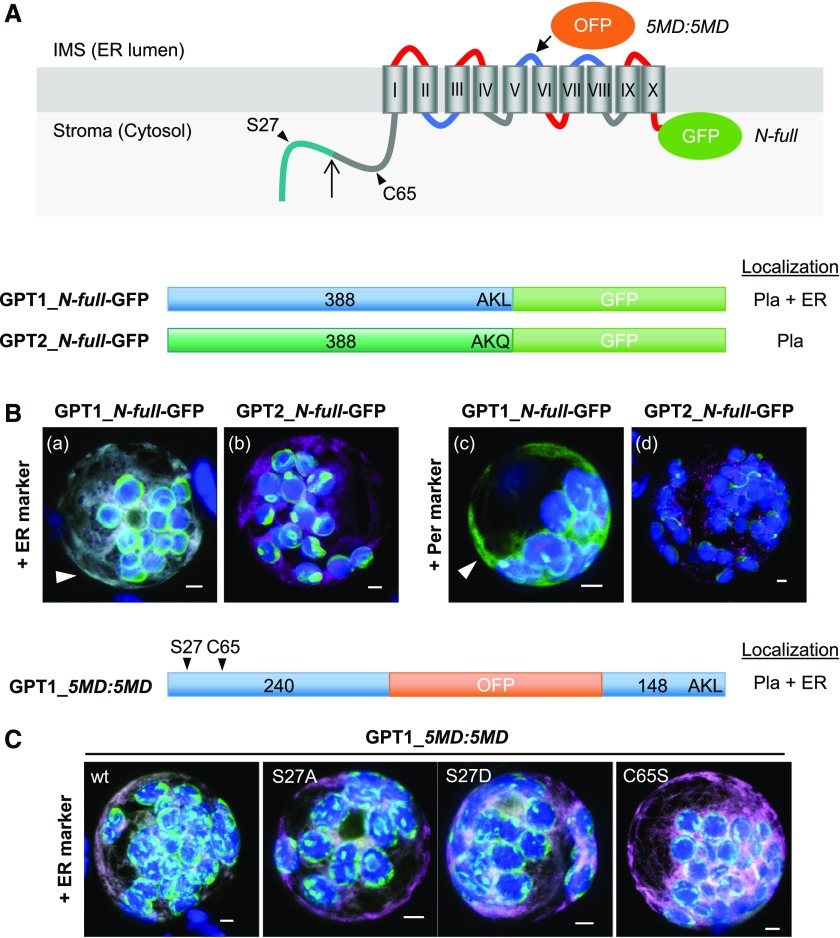
(A) Topology model of Arabidopsis G6P/phosphate translocator (GPT) isoforms with 10 membrane domains (MD) depicted as barrels (Roman numerals), connected by hinge regions (red, positive; blue, negative; and gray, neutral net charge), and both N-/C-terminal ends facing the stroma (Lee et al. 2017). Relevant positions are indicated: Plastidic TP (green), TP processing site (upward arrow), N-terminal amino acids potentially modified/regulatory in GPT1 (arrowheads), medial OFP insertion (5MD:5MD), and C-terminal GFP fusion (N-full). Pla, plastids.
(B) and (C) Localization of the depicted GPT-reporter fusions upon transient expression in Arabidopsis protoplasts (24- to 48-h post transfection).
(B) With free N terminus, GPT1 targets both plastids and the ER (a and c, arrowheads), but GPT2 only targets plastids (b and d). Scale bars = 3 μm.
(C) The medial GPT1_5MD:5MD construct (wt, wild type) was used to analyze potential effects of single amino acid changes in the N terminus: Ser27-to-Ala (S27A, abolishing phosphorylation), Ser27-to-Asp (S27D, phospho-mimic), and Cys65-to-Ser (C65S, precluding Ser modification). All images show maximal projections of ∼30 optical sections (shown as merged; for single-channel images, see Supplemental Figure 5). Candidate fusions are shown in green, ER marker (B, OFP-ER; C, GFP-ER) or peroxisome marker (OFP-PGL3_C-short) in magenta, and chlorophyll fluorescence in blue. Co-localization of green and magenta (or very close signals <200 nm) appear white in the merge of all channels. Scale bars = 3 μm.