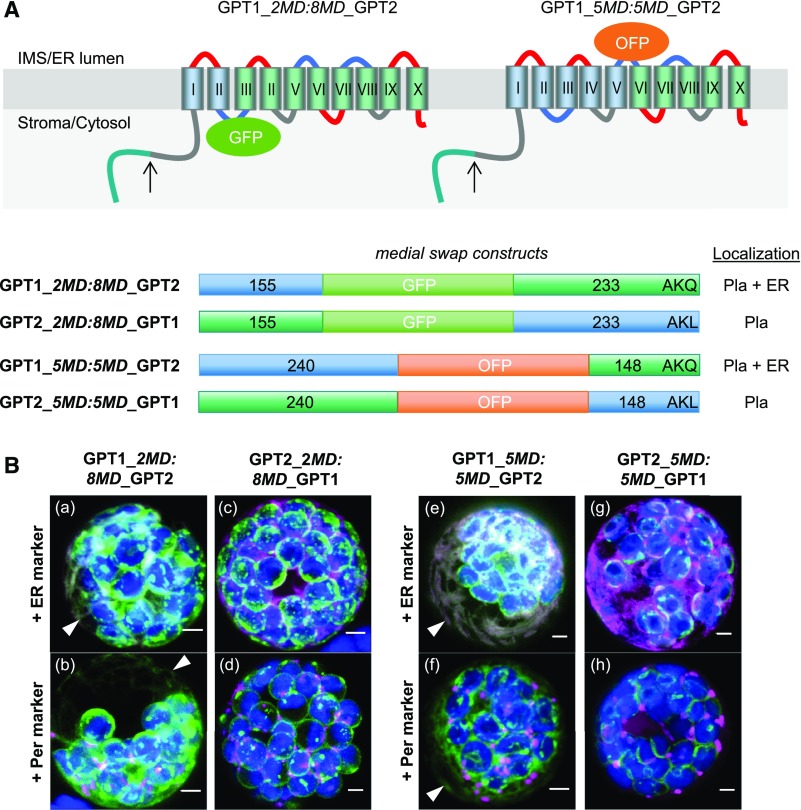
Figure 2.
Domain Swaps Demonstrate that the N Terminus of GPT1 Confers ER Targeting.
(A) Topology models of the GPT medial swap constructs showing the orientation of the inserted reporters: GFP facing the stroma/cytosol and OFP facing the IMS/lumen of the ER. Membrane domains (depicted as barrels, Roman numerals) of GPT1 in blue and of GPT2 in green. The upward arrows indicate TP cleavage sites (in the plastid stroma).
(B) Localization of the indicated medial swap constructs in Arabidopsis protoplasts (24- to 48-h post transfection). When headed by GPT1 (GPT1_2MD:8MD_GPT2 or GPT1_5MD:5MD_GPT2), both plastids and the ER (arrowheads) are labeled (a, b, e, and f); when headed by GPT2 (GPT2_2MD:8MD_GPT1 or GPT2_5MD:5MD_GPT1), only plastids are labeled (c, d, g, and h). All images show maximal projections of ∼30 optical sections (merged; for single-channel images, see Supplemental Figure 7). Candidate fusions are shown in green, ER marker (G/OFP-ER) or peroxisome marker (Per; G/OFP-PGL3_C-short) in magenta, and chlorophyll fluorescence in blue. Co-localization of green and magenta (and very close signals <200 nm) appear white in the merge of all channels. Scale bars = 3 μm.