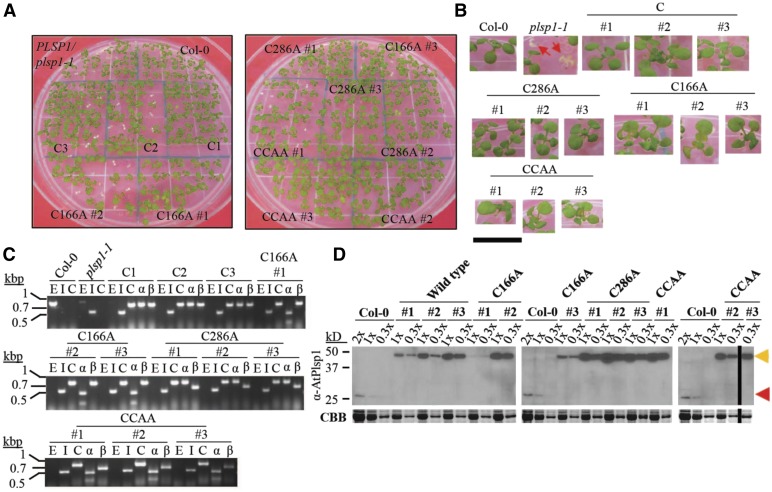
Figure 3.
Expression of Redox-Inactive Citrine-Plsp1 Variants Rescues the plsp1-1 Knockout Mutant.
(A) Images of 12-d-old seedlings growing on MS medium supplemented with 1% w/v Suc. C = CITRINE-PLSP1, C166A = CITRINE-PLSP1-C166A; C286A = CITRINE-PLSP1-C286A; CCAA = CITRINE-PLSP1-C166A/C286A.
(B) Close-up images of seedlings from (A). Red arrows indicate plsp1-1 null mutants. Scale bar = 1 cm.
(C) Results of genomic PCR experiment to confirm seedling genotypes. E = PLSP1; I = plsp1-1; C = CITRINE-PLSP1; α = PLSP1-C166A (PstI digestion of C); β = PLSP1-C286A (PvuII digestion of C).
(D) Expression of Citrine-Plsp1 proteins in isolated chloroplasts. Proteins from isolated chloroplasts were analyzed by SDS-PAGE and immunoblotting using the α-AtPlsp1antibody. RbcL bands on duplicate gels stained with Coomassie Brilliant Blue (CBB) are shown below as a loading control. The vertical black bar separates nonadjacent lanes from the same membrane. 1X =1 μg chlorophyll equivalent. Red and orange arrows indicate Plsp1 and Citrine-Plsp1, respectively.