Figure 6.
Association with PGRL1 Does Not Compensate for Lack of Cys in Plsp1.
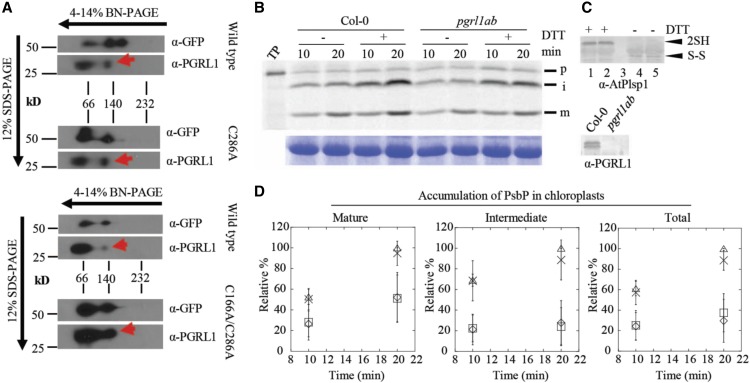
(A) 2D-Blue Native (BN)/SDS-PAGE analysis of Arabidopsis chloroplasts complemented with Citrine-Plsp1, Citrine-Plsp1-C286A, or Citrine-Plsp1-C166A/C286A. Total chloroplasts equivalent to 5 μg (left) or 7.5 μg chlorophyll were solubilized with 1.3% w/v n-dodecyl-β-D-maltoside and run on 4-14% Blue Native-PAGE gels. Lanes were then excised, heated at ∼80°C for 30 min in denaturing buffer (65 mM Tris-HCl pH 6.8; 3.3% SDS; 570 mM β-ME), and overlayed on 12% SDS-PAGE gels. After electrophoresis, proteins were blotted to polyvinylidene difluoride membranes. Membranes were cut at ∼37-kD and probed with the GFP antibody (top) or the PGRL1 antibody (bottom). Red arrows indicate the population of PGRL1 that comigrates with Citrine-Plsp1.
(B) Analysis of processing activity during in vitro import. Chloroplasts pretreated on ice for 30 min with or without 50 mM DTT were mixed with 35S-Met-prPsbP and MgATP (3 mM final) and were incubated in the light at 20°C for 10 or 20 min. Intact chloroplasts were reisolated as described in Figure 3B and analyzed by autoradiography (top panel). RbcL bands from the Coomassie Brilliant Blue–stained gels is shown as a loading control (bottom panel).
(C) Immunoblotting analysis of chloroplasts used for in vitro import experiments. For top panel, an aliquot of chloroplasts treated with or without 50 mM DTT before in vitro import were mixed with nonreducing (no β-ME) sample loading buffer and analyzed by SDS-PAGE and immunoblotting using the antibody against Arabidopsis Plsp1. ”2SH” and ”S-S” indicate the reduced and oxidized forms of Plsp1, respectively. 1 and 4 = Col-0; 2 and 5 = pgrl1ab; 3 = blank. For bottom panel, chloroplasts used for import experiments were analyzed by SDS-PAGE and immunoblotting using the PGRL1 antibody.
(D) Quantification of import products shown in (C). Bands were quantified relative to the corresponding band (S) in the Col-0 + DTT sample at 20 min, which was set as 100%. Data are expressed as the mean ± sd of three independent experiments. Squares, wild type - DTT; triangles, wild type + DTT; diamonds, pgrl1ab - DTT; X, pgrl1ab + DTT.