Applying an improved high-throughput gene-editing pipeline to functionally mapped candidates promises high-efficiency gene discovery by large-scale knowledge-informed mutagenesis.
Abstract
Maize (Zea mays) is one of the most important crops in the world. However, few agronomically important maize genes have been cloned and used for trait improvement, due to its complex genome and genetic architecture. Here, we integrated multiplexed CRISPR/Cas9-based high-throughput targeted mutagenesis with genetic mapping and genomic approaches to successfully target 743 candidate genes corresponding to traits relevant for agronomy and nutrition. After low-cost barcode-based deep sequencing, 412 edited sequences covering 118 genes were precisely identified from individuals showing clear phenotypic changes. The profiles of the associated gene-editing events were similar to those identified in human cell lines and consequently are predictable using an existing algorithm originally designed for human studies. We observed unexpected but frequent homology-directed repair through endogenous templates that was likely caused by spatial contact between distinct chromosomes. Based on the characterization and interpretation of gene function from several examples, we demonstrate that the integration of forward and reverse genetics via a targeted mutagenesis library promises rapid validation of important agronomic genes for crops with complex genomes. Beyond specific findings, this study also guides further optimization of high-throughput CRISPR experiments in plants.
Introduction
Global crop production will need to double by 2050 in order to feed the increasing world population. As one of the most important crops for food, feed, and fuel in agriculture, raising the yield of maize (Zea mays) will contribute to meeting our needs for food production beyond current projections (Ray et al., 2013). Most maize yield traits are quantitative, and cloning the causal genes and dissecting the underlying mechanisms affecting these traits are both key to continuous genetic improvement.
As a classical model system for genetic studies, hundreds of quantitative trait loci (QTL) for many traits have already been mapped in maize (Xiao et al., 2017; Liu and Yan, 2019). Nonetheless, the number of causal genes confirmed within these QTL regions is relatively small compared to rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana). Large-scale efforts aimed at genome-wide mutagenesis based on the random insertion of various elements in the genome (transposon, T-DNA, or the Tos17 retrotransposon) have been a key resource employed widely in rice and Arabidopsis over the last two decades (Jeon et al., 2000; Alonso et al., 2003; Wang et al., 2013). Although transposon tagging and mutagenesis by the Activator (Ac) and Dissociation (Ds) transposable elements (Cowperthwaite et al., 2002; Vollbrecht et al., 2010) and UniformMu (May et al., 2003; McCarty et al., 2005; Settles et al., 2007) or chemical mutagens such as ethyl-methanesulfonate (Lu et al., 2018) have all been used in maize, the exact identification of causal gene(s) among the tens or even hundreds of loci within a line that might have been mutated but are not responsible for the phenotype under question is still costly due to the complexity of the maize genome. The laborious and low-throughput nature of classical forward genetics approaches that rely on the segregation of the causal mutation(s) in a mapping population hinders the successful and rapid application of these resources in many plant species.
The RNA-guided CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) system represents a massive breakthrough both in terms of simplicity and efficiency (Cong et al., 2013; Mali et al., 2013) and has been extensively applied in plant genome editing since 2013 (Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013). Although more difficult to apply to plant species than to human cell lines (Yin et al., 2017), CRISPR/Cas9-based genome editing has recently been successfully applied to large-scale mutagenesis efforts in rice (Lu et al., 2017; Meng et al., 2017) and soybean (Bai et al., 2019). Due to its convenience, low cost, high specificity, and high-throughput scalability, CRISPR/Cas9-based editing therefore holds great promise for functional crop genomics. However, a proof-of concept study that demonstrates the feasibility and efficiency of such an approach is so far lacking for complex genomes such as maize.
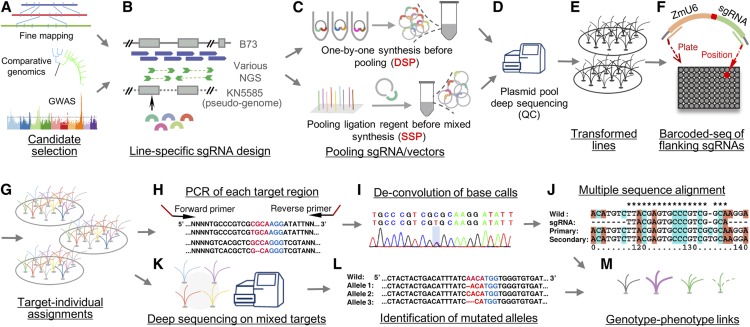
In this study, we report the development of a CRISPR/Cas9-based editing platform adapted to high-throughput gene targeting in maize, and its application in functional gene identification by integrating over 1000 candidate genes derived from genetic mapping and comparative genomic analysis (Figure 1). Through the use of state-of-the-art sequencing technologies and validation by Sanger sequencing, we established low-cost optimized and quality-controlled pipelines for each step, from the design of guide RNAs (sgRNAs) to the identification of targeted genes and edited sequences. Our study also expands on two key aspects that are critical during large-scale plant genome-editing research. First, general properties and insights for outcomes of plant genome editing were obtained and could serve as a reference for other crops. Second, knowledge-driven candidate genes were selected, and a large number of mutants were screened using lines from T1 or follow-up generations. Our results indicate that the integration of high-throughput gene-editing and forward-genetic approaches has great potential in rapid functional gene cloning and validation.
Figure 1.
Pipeline of High-Throughput Genome-Editing Design.
(A) Candidates selected from QTL fine mapping, genome-wide association mapping studies (GWAS), and comparative genomics.
(B) Line-specific sgRNA filtering based on assembled pseudo-genome of the receptor line KN5585.
(C) Different vector construction approaches of double sgRNA pool (DSP) and single sgRNA pool (SSP).
(D) Measuring the coverage and uniformity during plasmid pool by deep-sequencing.
(E) to (G) Transformation and assignment of targets to each T0 individual by barcode-based sequencing.
(H) to (J) Identification of mutant sequences by Sanger sequencing.
(K) and (L) Identification of mutant sequences by Capture-based deep-sequencing.
(M) Measuring phenotype changes and identification of functional genes.
RESULTS
Establishment of CRISPR/Cas9-Based Batch Targeting System
Based on existing and tested vectors for maize (Li et al., 2017) and rice (Lu et al., 2017) transformation, three vectors were optimized to allow one-step construction via overlapping PCR combining homologous recombination or T4 DNA ligase ligation (Supplemental Figure 1; see Methods). These vectors are suitable for pooled CRISPR/Cas9-based knockout for individual sgRNAs or paired sgRNAs in each plasmid.
For all three vector types (Supplemental Figure 1), we used the maize inbred line KN5585 for Agrobacterium tumefaciens-mediated transformation of immature embryos, with an average 14% transformation efficiency (Supplemental Table 1). To explore the gene-targeting efficiency of our constructs, we designed four sgRNAs within a single plasmid to target the ZmPLA1 (PHOSPHOLIPASE A; Liu et al., 2017a), resulting in a mutation rate ranging from 79% (23/29) to 83% (24/29) in the T0 generation (Supplemental Figure 2). This high-targeting frequency is consistent with a previous study (51 to 91%; Li et al., 2017) and may be a consequence of using a maize endogenous RNA polymerase III promoter to drive the expression of the sgRNA (Qi et al., 2018). Even though the relatively low transformation efficiency in maize presents a massive challenge, the high targeting efficiencies of these vectors rendered subsequent experiments possible.
Choice of Candidate Genes for Batch Editing
A total of 1244 candidate genes were collected for pooled knockout experiments and functional validation. The candidates were divided into two sets. Set no. 1 included 98 genes that had been either (1) fine-mapped to regions with one to a few candidate genes by linkage mapping or (2) derived from comparative genomics, as each individual gene showed a high probability of being associated with various traits. Set no. 2 was made up of 1181 genes, mainly from 70 mapped QTL regions corresponding to 27 agronomically relevant traits and including 35 genes that overlapped with those from set no. 1 (see Methods; Supplemental Figure 3). These candidate genes served as a springboard for building the batch editing pipeline. This study also intended to establish a preliminary targeted mutant library for maize functional genomic studies.
Since the KN5585 line originates from the tropics, its genome differs significantly from the B73 reference genome. We therefore established a new pseudoreference by deep sequencing of genomic DNA (to ∼60× coverage) and RNA samples collected from seven diverse tissues. Assembled contigs were used for genotype-specific sgRNA design (see Methods; Figure 1B). sgRNAs obtained by this method were confirmed by Sanger sequencing on all set no. 1 candidates, ensuring high reliability of sgRNA design. Double sgRNAs in one vector were designed primarily for set no. 1 genes (double-sgRNA pool, DSP), with the expectation that this would increase the probability of obtaining knockout lines. Individual sgRNAs per vector were used for set no. 2 genes (single-sgRNA pool, SSP). These two sets were used separately, leading to a total of 1290 vectors consisting of 1368 sgRNAs for 1244 genes.
High Uniformity and Coverage of sgRNAs during Pooled Construction and Transformation
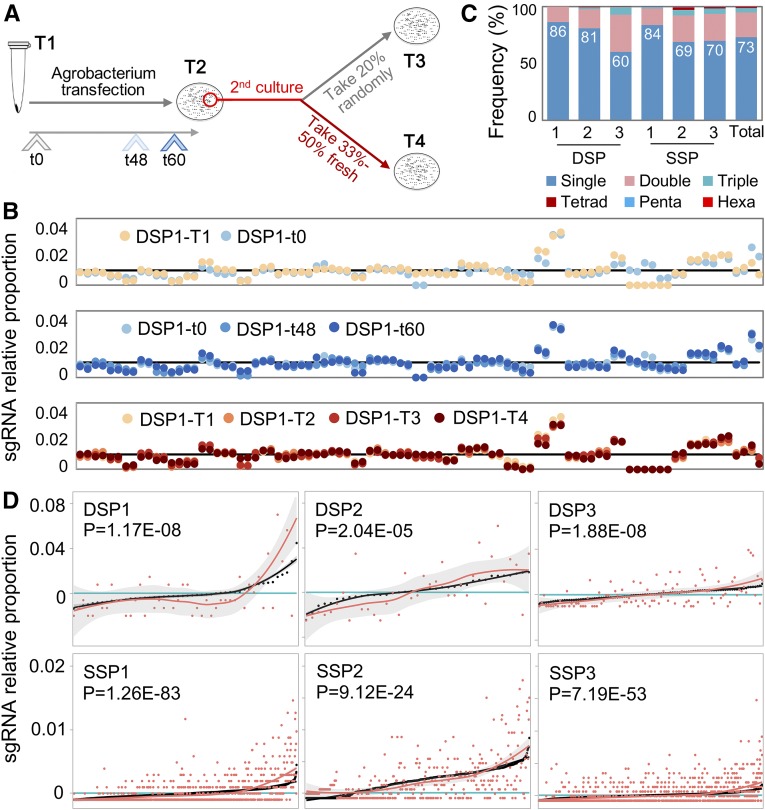
Coverage and uniformity are two key factors during pooled transformations, so that all cloned vectors are represented within pools. Since only the spacer sequences (e.g., 20 bp) of sgRNAs differed between vectors, primers from flanking sequences were used to amplify these sequences for next-generation sequencing (NGS) in order to evaluate the relative presence of different sgRNAs. No significant differences were observed between the two pooling strategies, that is either pooling after construction for the DSP gene set (mixing the vectors separately) or pooling after ligation for the SSP gene set (mixing ligation reagents first, followed by pooled construction). Indeed, both had acceptable uniformity and coverage for sgRNA distribution. Nevertheless, pooling after ligation was easier to implement. The uniformity and high coverage for sgRNA distribution was also stable following different culture periods and after Agrobacterium transfection (Figures 2A and 2B).
Figure 2.
High Coverage and Uniformity from Plasmid Pool to T0 Individuals.
(A) and (B) Plasmid sequencing in quality-control process (A), results of measuring the coverage and uniformity of sgRNA amount (B). T1, Primary plasmid pool before Agrobacterium transfection, at t0. T2, Plasmid pool extracted from the first Agrobacterium colonies. T3, Plasmid pool randomly extracted from 20% of colonies of second Agrobacterium transfections. T4, Plasmid pool specifically taken from 33 to 50% of fresh and more vigorous colonies of second Agrobacterium transfection for further embryo transformation. t0, The primary plasmid pool before Agrobacterium transfection; t48/t60, 48 h or 60 h culture on solid medium after Agrobacterium transfection. The sgRNAs are ordered along the x axis based on their ID number. The "DSP1-T1" and "DSP1-t0" in the top panel here were equivalent to technical repeats.
(C) Ratio of coinfection events in six batches (three SSPs and three DSPs) and total.
(D) Correlation of sgRNA relative amount between plasmid pool (black) and T0 individuals (red). Proportion lines were smoothed. All sgRNAs along the x axis were sorted according to their relative proportion in the plasmid pool.
The coverage of pooled sgRNAs was high, 98% on average. Only a few sgRNAs could not be detected at any given stage. This may be caused by sequencing bias, since undetected sequences usually could be found at other stages. For example, 52 of the 1181 gRNAs from SSP were not detected before the transformation but were subsequently identified in T0 plants. Together, these results implied that coverage was uniform and sufficient to construct a mutant library.
A Barcode-Based NGS Approach Reveals the Uniformity and Coverage of sgRNAs in T0 Plants
Six CRISPR libraries of sgRNAs were separately transformed into immature embryos via cocultivation with Agrobacterium tumefaciens, and a total of 4356 T0 seedlings resistant to the herbicide glyphosate were transplanted (Table 1). DNA from leaves of each T0 seedling was sampled at least in duplicate, and sgRNA-specific PCR followed by barcode-based deep sequencing was performed to identify the corresponding target(s) within each plant (Figure 1D; Supplemental Figure 4). Care was taken to ensure high reliability of target determination (see Methods; Supplemental Figure 5). In total, 3695 (or 85%) of T0 plants were reliably assigned to 778 vectors corresponding to 743 target genes and used for further analysis, while unconfirmed plants were verified in additional experiments. Most positive T0 plants (2704, or 73.2%) carried single event, while double and triple coinfections were found in 21.5% and 3.8% of cases (Figure 2C), respectively.
Table 1. Statistics of the Genome-Editing Experiments.
| Pool | Batch | sgRNAs | Vector No. (Vn)a | V′n in Plasmidb | T0 No. | Assigned T0 (Pn)c | V′n in T0 Plantsd | Genotyped Linese | Edited Lines |
|---|---|---|---|---|---|---|---|---|---|
| DSP | DSP1 | 90 | 49 | 48 | 157 | 125 | 38 | 95 | 79 |
| DSP2 | 78 | 40 | 37 | 342 | 296 | 34 | 263 | 224 | |
| DSP3 | 191 | 100 | 98 | 387 | 379 | 75 | 173 | 146 | |
| DSP | 191 | 104 | 103 | 886 | 800 | 93 | 531 | 449 | |
| SSP | SSP1 | 959 | 959 | 936 | 940 | 860 | 340 | – | – |
| SSP2 | 1186 | 1186 | 320 | 1374 | 1016 | 257 | – | – | |
| SSP3 | 1186 | 1186 | 1173 | 1156 | 1019 | 466 | – | – | |
| SSP | 1186 | 1186 | 1178 | 3470 | 2895 | 685 | 1,290 | 693 | |
| Total | 1368 | 1290 | 1281 | 4356 | 3695 | 778 | – | – |
Dashes indicate no data. Bold text represents the sum of each pool and the total of them.
Total vector number (Vn) pooled in this study.
Observed vector number (V′n) in plasmid pools.
T0 individuals successfully assigned to linked targets.
Vector number covered by those successfully assigned T0 individuals.
The number genotyped for DSP is indicated by the total T0 lines. The number of T1 lines with phenotypic change were selected for SSP genotyping (thus it is inappropriate and not used for estimation of general mutant ratio).
The number of T0 plants isolated for a given sgRNA was positively correlated (P < 2.0E-5) with the amount of each sgRNA in the plasmid pool, although differences were slightly magnified in the transgenic lines (Figure 2D), implying that a balanced vector pool is necessary to obtain a balanced maize mutant library. On average, 4.3 T0 individuals were obtained for each target sgRNA (Table 1). We used a simulation analysis to model that 4 to 10 T0 plants (relative to gene/vector number) were required to cover at least 98% of the chosen candidate genes (see Methods). Interestingly, our simulation analysis suggested that the number of mixed vectors in each batch should be over 50 in order to avoid large deviations from the expected coverage (Supplemental Figure 6).
Efficient Identification of Sequence Variation in Edited Plants
Identification of induced sequence variants with high sensitivity and accuracy remains a challenge for high-throughput experiments. Using Sanger sequencing, we found 449 (out of a total of 531, or ∼85%) T0 individuals from the DSP with mutations at target loci, and 118 (26%) had large deletions between two sgRNAs. Sanger sequencing was inadequate for accurate variant identification, especially for individuals with multiple variants, and was also time consuming and labor intensive when many lines and/or genes were analyzed.
We therefore developed an improved method based on the MassARRAY system, which is usually used for genotyping known variants (Ellis and Ong, 2017), with sequential primer combinations to infer the as-yet-unknown mutated alleles. This method was particularly suitable for efficient medium-scale (20 to 50) gene identifications (Supplemental Figures 7 and 8; Supplemental Table 2) and was used in a single experiment to successfully identify 24 lines with exact mutations among 30 randomly selected T0 individuals from the SSP experiments. These results were consistent with Sanger sequencing. The observed mutation rate in the SSP was estimated to be around 80% (24 of 30), slightly lower than that of DSP (83%∼85%).
In order to scale up the method to allow for high-resolution detection of induced mutations to many genes and to render the method capable of estimating allele-specific mutation efficiency, we turned to target-region capture-based sequencing (TRC-seq; see Methods). We designed 113 primers for 106 genes to capture regions flanking sgRNA target sites from T1 lines with obvious morphological changes. Since we had already identified their respective individual target genes during the T0 generation, 20 to 25 individuals with different targets could be combined into a batch for TRC-seq without compromising on sensitivity. A total of 1208 unique T1 lines from 60 pools were assayed by this method, of which 656 were also characterized by Sanger sequencing. We used the improved biologically informed alignment algorithm CRISPResso2 (Clement et al., 2019) for deconvolution of edited alleles from deep-sequencing data. Mutated alleles identified by TRC-seq included all the homozygous mutations that we had identified by Sanger sequencing, indicating its high sensitivity.
While a median of 81% of edited genes identified by TRC-seq was consistent with previous target assignment, the remaining 19% of mutations, from 19 genes, were newly identified, compared with previously assigned individuals/targets. These results demonstrated (1) the highly reliable but conservative target assignment and (2) the superior efficacy of the TRC-seq method in mutation identification. Even though CRISPResso2 has multiple advantages in the identification of mutant alleles, it also had a propensity for false-negative discovery, since a large number (130 of 292, or 39%) of lines, covering a total of 32 genes, were identified as homologous alleles exclusively by the Sanger method. To explore the contribution of rigorous filtering and alignment procedures, a standard variant calling pipeline followed by global mapping of short reads to the pseudogenome was additionally integrated in order to detect mutant alleles (see Methods). With an acceptable reliability of only three lines (out of 166, ∼2%) differing from the overlapped homologs called by Sanger method, this method remedied nearly 40% (51 of 130) of the CRISPResso2 false negatives. However, 27% (79 of 292) false-negative discoveries (compared to Sanger sequencing) still remained, possibly caused by the biased mixing of individuals and asymmetrical capture during deep sequencing.
Pattern and Predictability of Mutations Generated by Editing
Considering the complementary ways in which our different methods addressed mosaicism (described below in detail), the mutations identified from SSP and DSP pools using Sanger sequencing and TRC-seq were merged for further analysis. A total of 326 unique mutant sequences in 109 genes corresponding to 135 individual sgRNAs were collected. An additional 86 nonredundant structural variants between paired sgRNAs of 53 genes were also identified (Supplemental Data Set 1), providing a representative resource to understand the genome-wide distribution of editing in maize.
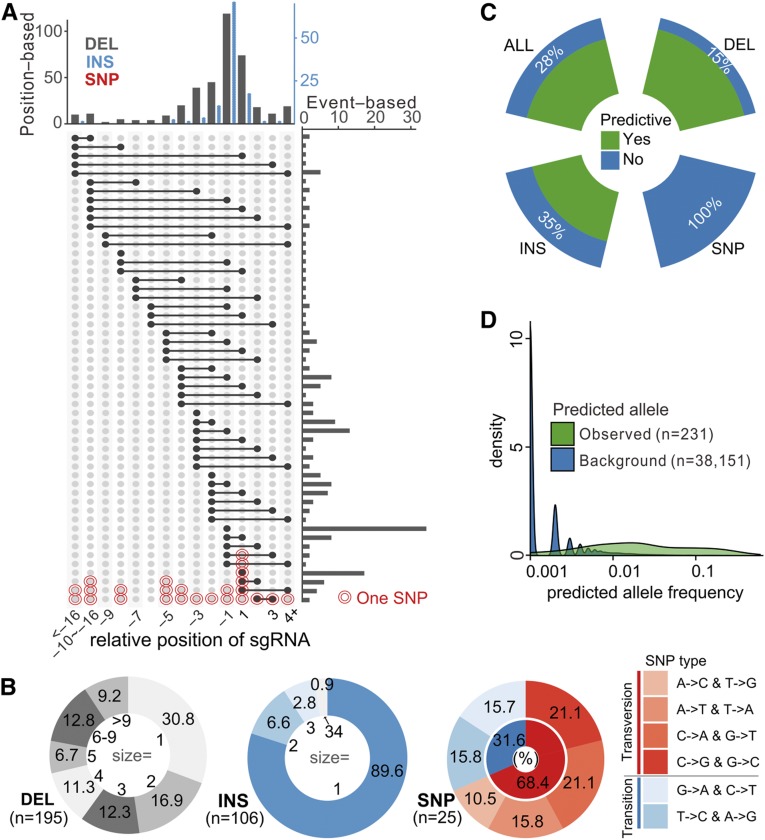
For the individual target mutated sequences, most (60%) were deletions (DELs) of 1 bp to 65 bp, with a median of 3 bp. Breakpoints were enriched within a 4-bp window 3 to 6 bp upstream of the NGG PAM (protospacer-adjacent motif) sequence. Insertion-type (INS) mutants accounted for nearly one-third (32.5%), with 90% being single bp insertions and usually occurring within the predicted nuclease cleavage site (three to four nucleotides upstream of the PAM; Figure 3A). Most of the remaining mutations (8%) were single nucleotide polymorphisms (SNPs), transversions being twice as frequent as transitions. Individual sgRNAs sometimes produced large deletions or insertions. In contrast, when using paired sgRNAs, we often observed structural variants between the target sites, with deletions being the most frequent (91%; Supplemental Figure 9A). For genes targeted with two sgRNAs, whether a large deletion between the two sgRNAs or a small insersion or deletion (InDel) at each individually sgRNA target site was induced could not be predicted (Supplemental Figure 9B), although the distance between paired sgRNAs was found to slightly affect the outcomes (Supplemental Figures 9C and 9D).
Figure 3.
Mutation Pattern and Predictability of Variants Generated by Genome Editing.
(A) Allele size and position distribution based on all individual events. Position-based, distribution along relative position on sgRNA; event-based, distribution of individual events. The position along sgRNA (x axis) is relative to predicted nuclease cleavage site, while +1 and −1 indicate the nucleotides 3 to 4 bp upstream of the PAM. INS, Insertion; DEL, deletion.
(B) Distribution of mutant outcome sizes (in bp) and diversity for different mutant classes.
(C) Ratio of real observed alleles that are being predicted by only flanking sequences, classified by mutant types (DEL, INS, SNP). ALL corresponds to all mutant types added.
(D) Algorithm-mediated prediction of mutant outcomes based on flanking sequences. The set of alleles observed in real cases display significantly higher predicted frequency compared with all predicted outcomes (background).
Recent studies suggest high predictability of genome editing in human cell lines (Shou et al., 2018; Chakrabarti et al., 2019), and an algorithm to predict mutational outcomes using only flanking DNA sequences has been described (Allen et al., 2018). Interestingly, even though the algorithm was refined using human cell line data, it was able to predict the outcome of 72% of the observed alleles in this study, and this increased to 85% for DELs (Figure 3C). Furthermore, the algorithm estimated allele frequencies for true observed variants much better than background (P = 2.3E-16; Figure 3D), suggesting that primary alleles were readily captured. Despite the fact that many of the mutants not predicted by the algorithm were large (for example, 24% of such nonpredicted DELs were longer than 10 bp) and the presence of cell-line-dependent bias (Allen et al., 2018), the predictions developed from human data are therefore largely transferable to plants. Even though plants have unique mechanisms for repair of double-strand breaks (Spampinato, 2017) and somewhat different mutation signatures are observed between animals and plants (Bortesi et al., 2016), our study provides the justification to apply mutant allele prediction in advance of sgRNA design to guide precise editing in plants.
We next used a tree-based Random Forest algorithm to test the effect of sgRNA sequences in predicting the outcomes produced in this study. Given the limited data size, the general accuracy on classifying the mutant types (INS, DEL, or SNP) from sgRNA sequences was low (Supplemental Figure 10). To ask what additional factors beyond sgRNAs and their flanking DNA sequences might affect editing outcomes, we also considered the expression patterns of the candidate genes as an additional explanatory variable (Supplemental Figure 10A). Interestingly, the expression variability of target genes along diverse tissues affected the size of InDel events and the position of DELs, as higher expression variability was associated with smaller mutations that were more proximal to the predicted nuclease cleavage site (Supplemental Figures 10D and 10G). SNPs in target genes with higher expression in the shoot apical meristem also appeared to be more proximal to the predicted nuclease cleavage region (Supplemental Figure 10F and 10G). Previous studies also found that chromatin states and active transcription affect Cas9 binding (Verkuijl and Rots, 2019) and editing mutant profiles (Chakrabarti et al., 2019) and thus further exploration on how expression changes influence mutational outcomes could lead to improved predictability.
Homology-Directed Repair with Endogenous Templates as a Means of Mutant Generation
Programmable nucleases introduce DNA double-strand breaks at user-defined target sites and thus engage the inherent repair systems such as error-prone nonhomologous end joining or, in the presence of a DNA template, homology-directed repair (HDR). Among the mutants identified from TRC-seq of SSP T1 lines, we identified two clear cases of HDR that used interchromosomal endogenous templates (Supplemental Figure 11). Given the total of 154 mutated InDels covering 63 genes, these two cases accounted for 1.3% and 3.2% of total mutations and genes, respectively, suggesting a much higher frequency than previous reports in plants (Puchta, 1999; Ayar et al., 2013). Evidence for the hypothesis that nonhomologous end joining repair occurred sequentially after initial cleavage, resulting in HDR, was also observed (Supplemental Figure 11B). The estimated mutant frequencies caused by HDR were 1% and 20% for these two genes, respectively. These ratios were comparable to studies that improved HDR efficiency using exogenous templates in plants (Gil-Humanes et al., 2017; Wang et al., 2017a; Li et al., 2019b). An improved genome assembly of the maize transformation recipient line used here (KN5585) will improve the detection of more endogenous HDR events.
The targets and corresponding templates for the two documented cases of HDR were homologues with highly correlated expression patterns (Supplemental Figure 11C). Interestingly, for one case, the chromatin bearing the homologous template and the target gene were shown to come in close proximity to each other, although they are located on different chromosomes (Supplemental Figures 11C and 11D; Peng et al., 2019), suggesting that higher-order chromatin structure contributes to the high frequency of endogenous HDR. This finding supports the hypothesis that low frequency of precise gene replacement through HDR in plants might be due to an inefficient targeting of exogenous templates, as opposed to a difference in endogenous repair mechanisms compared to mammals (Schuermann et al., 2005; Lieberman-Lazarovich and Levy, 2011; Fauser et al., 2012). Further study of these endogenous HDR events might provide clues toward optimizing HDR efficiency, and thus improving the efficiency of precise introduction of specific variants.
Rare Off-Target versus Common Mosaic Mutations
Consistent with previous studies that found rare off-target events in plants when using CRISPR/Cas9 (Tang et al., 2018; Li et al., 2019a), we identified only 10 InDels among a total of 39,328 potential off-target genes via whole-exome sequencing in 19 mixed T1 blocks covering 25 mutated genes (see Methods). Thus, off-target effects will likely have only a small effect on plant editing, at least under our conditions. By contrast, mosaic mutations were observed widely in this study. Evidence from SSP T1 lines indicated that (1) most heterozygous alleles called from Sanger sequencing were biallelic, and only 1.4% (2 of 148) included one wild-type copy; (2) only 46% of variants from capture sequencing (TRC-seq) were matched to one of the heterozygous alleles detected by Sanger sequencing, while the remaining 54% were different; (3) different homozygous mutations were observed among T1 individuals from the same self-crossed T0 ear; and (4) base calls with Sanger sequencing of 41 lines were completely impossible to interpret, most likely a coexistence of more than two alleles at a given locus. Such chimeras can impair mutant characterization and inference of any genotype-phenotype links. For example, even though a large deletion was identified for one flowering-time candidate in a T0 event, no mutation was found in a large number of derived T1 lines. This finding calls for higher scrutiny not only for mutation identification but also for further validation of genotype-phenotype association.
Knowledge-Driven Gene Editing Accelerates the Exploration of Gene Function
The edited lines provided reliable evidence in causal gene validations for selected candidates that were previously fine-mapped to individual genes (DSP set). For example, they provided confirmation for the validation of ZmDXS2 (1-DEOXY-D-XYLULOSE-5-PHOSPHATE SYNTHASE 2; GRMZM2G493395) in affecting kernel color and carotenoid contents (Fang et al., 2020). Although lines carrying only 32% of the mutated genes were planted, some phenotypes were found to be consistent with predictions from forward genetics or comparative genomics, even though a large fraction of candidates (∼40%) from the SSP set were not mutated. We planted 639 T1 families from 445 SSP T0 events covering 246 genes and observed 119 T1 families representing 107 genes with significant morphological phenotypes. Importantly, we observed 13 genes showing altered phenotypes that were consistent with their QTL mapping predictions. Each QTL interval covers multiple genes, only one or very few of which might be expected to be responsible for the underlying phenotypes. We may have therefore missed the causal locus when designing our gene-editing constructs.
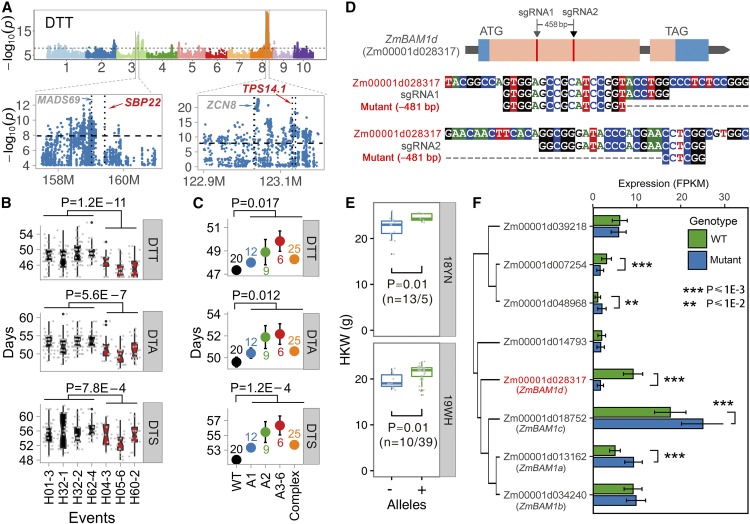
In addition, the mutants we generated are also valuable to identify new gene functions within classical QTL intervals. Taking flowering time as an example, the maize anti-florigen gene ZEA CENTRORADIALIS 8 (ZCN8) is usually assumed to be the causal locus behind the largest effect QTL on chromosome 8 that was mapped in various maize populations (Buckler et al., 2009; Coles et al., 2010; Liu et al., 2016; Guo et al., 2018), given this gene’s role in flowering regulation (Meng et al., 2011; Lazakis et al., 2011). However, this QTL region covers over 2 Mbps (Figure 4A) and suggests that variation in genes outside of ZCN8 might participate in the underlying QTL. Interestingly, mutants in ZmTPS14.1 (TREHALOSE-6-PHOSPHATE SYNTHASE 1, GRMZM2G068943, ∼100 kbp downstream of ZCN8) also displayed a significant delay in flowering time (Figure 4B; Supplemental Figure 12A and 12B), consistent with a previous study in Arabidopsis (Wahl et al., 2013). Another flowering time QTL on chromosome 3 was also associated with ear height (Figure 4A; Supplemental Figure 12A), and while the MADS-box transcription factor ZmMADS69 (GRMZM2G171650) located within this region was recently validated as a gene underlying flowering time regulation in maize (Liang et al., 2019), we obtained many mutated alleles of SQUAMOSA promoter BINDING PROTEIN gene ZmSBP22 (GRMZM5G878561, ∼370 kbp upstream of ZmMADS69) in this study, and all showed late flowering (Figure 4C; Supplemental Figure 12C and 12D). These findings raise the possibility that multiple causal genes might map to the same QTL regions and might contribute, alone or in combination, to the underlying phenotype, which is not easily addressed by routine genetic mapping analyses.
Figure 4.
Applied Targeted Mutagenesis for the Validation of Gene Function.
(A) Two large-effect flowering time QTLs for days to tasseling (DTT) identified by genome-wide association mapping studies (GWAS) and targeted by genome editing. Corresponding results for days to anthesis (DTA) and days to silking (DTS) are shown in Supplemental Figure 12A. Both QTL intervals include well-known causal genes (shown in gray), while novel genes identified in this study are shown in red. Significant flowering time differences are seen for Zmtps14.1 (B) and Zmsbp22 (C). Phenotypic values from wild-type lines are indicated in black, and all colors show mutant lines. Trait values for Zmtps14.1 and Zmsbp22 were measured as Jilin (northeast China, temperate climate) and Hainan (south China, tropical climate), respectively. H04-3, H05-6, and H60-2 refer to three independent T2 populations carrying the same allele. Corresponding edited alleles along the x axis are detailed in Supplemental Figures 12B and 12C.
(D) to (F) Gene redundancy from homologous genes can skew the results of a targeted gene.
(D) Two sgRNAs were designed to target the first exon of ZmBAM1d and caused a large deletion between sgRNAs. Both sgRNAs were specific for ZmBAM1d without affecting homologous genes.
(E) Selfing T3 edited lines carrying the deletion were used to measure kernel weight (HKW); only a marginal phenotypic difference was seen at both Yunan (year 2018, labeled as 18YN) and Wuhan (year 2019, labeled as 19WH). Overexpression lines have significantly higher HKW, and near-isogenic lines show significant differences in HKW (Yang et al., 2019), leading to the expectation that Zmbam1d edited lines would demonstrate smaller HKW.
(F) Expression of Zmbam1d and its homologous genes across three edited lines and corresponding wild-type segregants. Two of the three close homologs show higher expression that might compensate for the loss of Zmbam1d.
A loss-of-function allele induced by CRISPR-mediated gene editing may have different phenotypes from a subtle difference in protein function resulting from the underlying variation between naturally occurring alleles at a QTL. For example, GRMZM2G331652 (a gene encoding an aminotransferase-like protein) was located within a plant height QTL interval but falls outside of a small effect flowering QTL interval on chromosome 1 (Supplemental Figure 13A). Interestingly, in addition to the expected plant height changes, mutants in this candidate were also characterized by flowering time differences and varied responses to day length (Supplemental Figure 13D). Finally, as was our hope, we obtained lines with a large number of unexpected phenotypic changes, including traits not previously studied (Supplemental Figure 14) affecting plant size and morphology, reproductive structures, or susceptibility to disease, demonstrating that our library of edited genes provides an unprecedented resource for further detailed functional genomics.
The mutant library may also refute standing hypotheses of gene function and together would promote a new perspective on underlying regulatory mechanisms. An interesting case was for the BARELY ANY MERISTEM 1d gene ZmBAM1d (GRMZM2G043584), which was previously found to affect kernel weight and validated by results from a NIL population and overexpression (Yang et al., 2019). However, our CRISPR/Cas9-edited lines had no obvious phenotypic differences compared with the parental line (Figures 4D and 4E). RNA sequencing revealed the up-regulation of two BAM1d homologues as potential cause for the lack of visible phenotypes (Figure 4F), suggesting that a compensatory mechanism might be the reason for the lack of trait changes in the genome-edited lines. While gene redundancy is widely recognized as an obstacle to identifying gene function in plants, gene editing can be multiplexed to address this issue.
DISCUSSION
The CRISPR/Cas9 system is a simple, effective method for generating targeted mutations, and its capacity for high throughput has fueled its popularity in large-scale mutagenesis libraries, first in animals (Peng et al., 2015; Shalem et al., 2015) and now in plant systems (Lu et al., 2017; Meng et al., 2017; Bai et al., 2019). These benefits make the CRISPR-based system far outweigh other classical plant mutant libraries generated by transposon insertion of chemical mutagens. Here, we provide a practical workflow for high-throughput genome editing in maize, with optimized bioinformatic analysis, that should circumvent problems associated with its large and complex genome and difficulty of transformation (Figure 1). We anticipate that our approach is also applicable to other species. In contrast to human cell line screening, large-scale exploration of mutants and corresponding phenotypic analysis in plants is challenging, mainly due to the lower associated throughput, labor-intensive phenotyping and environmental impact during phenotyping in the field. This is especially true when large field trials are needed to detect small quantitative changes and when different environmental conditions (stress, nutrition) may reveal additional phenotypes. However, this will likely be addressed in the future via innovations in high-throughput phenotyping methods. As technologies for genome editing rapidly advance, emerging toolkits will be integrated into such future experiments. While recent studies offer high transformation efficiency for a wide variety of maize genotypes (Lowe et al., 2016, 2018; Jones et al., 2019), new methods in sgRNA delivery by viral vectors (Wang et al., 2017a) or by clay nanosheets (Mitter et al., 2017) that avoid the time-consuming tissue culture may be critical in accelerating functional genomics.
Here, we explored the CRISPR-Cas mutational profiles of a representative set of genes. The patterns of repair outcomes in our study were in line with those seen in human cell lines (Allen et al., 2018). Genome-editing events in the form of deletions and insertions largely dominated over SNPs, and the size of deletions varied more widely than that of insertions. This similarity allowed a good predictability of mutational outcomes in maize using an algorithm refined for human cell lines using only local sequences as input. Our findings suggest that the mechanisms of both Cas9-induced double-strand break and subsequent DNA repair are highly conserved between humans and plants. The prediction algorithm can thus be incorporated with sgRNA design and variant effect prediction to help prioritize sgRNAs based on expected mutant alleles and/or expected effect (such as frameshift or missense) on the target gene. This is important, since the precise introduction of given variants through repair of exogenous templates is still difficult, and a prescreening step of all possible sgRNAs for accurate prediction followed by screening of a smaller pool of mutated descendants is more tractable. Furthermore, this study provides evidence that the chromatin state (open chromatin being associated with higher expression and accessibility) at a targeted gene may have an impact on editing efficiency and on mutational outcomes, which can be further integrated for prediction improvement.
Cloning and validating genes affecting important agronomic traits remains key to crop genetic improvement, especially when implemented to target multiple traits each with multiple candidate regions; it is essential to meet future food demand. Mutants created by CRISPR/Cas9 are highly valuable in functional genomics, especially when used in a multiplex fashion. As screening phenotypic changes in a genome-wide mutant library is challenging in crops, access to candidate regions for corresponding traits identified by forward-genetic approaches is thus highly valuable. In this study, we integrated candidates from genotype-phenotype associations and CRISPR/Cas9 early on in our pipeline, and we provide a practical roadmap for the rapid detection of gene function through an informed mutagenesis library. In addition to the validation of high-confidence candidates, the approach may allow us to rule out other predicted candidates. At the same time, other mutants derived from the present design will be a valuable resource in functional gene discovery. Since candidates from natural variation have greater utility in crop improvement, such knowledge-driven targeted mutagenesis based on QTLs, pathways, and gene families will dramatically improve future studies. We anticipate that all candidate genes from a given QTL region can thus be mutated simultaneously in one implementation. Of course, complete gene loss of function alleles induced by genome editing may display drastic phenotypes that go beyond the range conferred by natural alleles: these validation experiments should be interpreted carefully. The heritable transmission ratio is also an important issue to test genotype-to-phenotype links but could not be explored in this study since the T0 and T1 populations were descended from unrelated individuals. However, previous studies in maize indicate that CRISPR/Cas9-derived mutations in T0 individuals were stably transmitted to the next generation (Zhu et al., 2016; Li et al., 2017), one of which used the same vector we did (Li et al., 2017). We also found that off-target mutations may not be common in plants, although editing at nontarget homologous sequences deserves attention and stresses the need for high-quality genomes of the parent lines.
The knowledge-informed mutagenesis design we present here is not only helpful in accelerating gene discovery; it will also be valuable to characterize the effects of specific genes or alleles, to study regulation mechanisms, to evaluate pleiotropic effects, and to create novel useful haplotypes. A multitude of CRISPR-derived alleles, with effects other than complete loss of function (a nonexhaustive list includes knockin, knock-down, or knock-up at specific developmental stages, base editing, or modifying epigenomic, transcriptional, or posttranscriptional processes) can be flexibly incorporated into fine tuning of regulatory networks (Chen et al., 2019; Hua et al., 2019; Zhang et al., 2019). The knowledge and materials available here therefore represent important tools in the acceleration of high-precision crop breeding (Fernie and Yan, 2019).
METHODS
Collection of Candidate Genes
The candidates selected for this study were from multiple sources:
(1) Genes that have been fine-mapped using various recombinant inbred line populations. Most traits mapped to single genes, and a few mapped to intervals with several (less than five) genes. Additional genes included four related to tocopherol content, four to carotenoid content/composition, three to kernel dehydration rate, three to maize (Zea mays) leaf blight susceptibility, three related to ear yield, and one to tassel length.
(2) 19 genes from the CCT (CONSTANS, CO-like, and TOC1) domain-containing family with high potential for affecting maize flowering time (14 of which were orthologs from rice [Oryza sativa] and Arabidopsis [Arabidopsis thaliana]), located within QTLs for flowering time identified by genome-wide association mapping studies in a recently developed population (Liu et al., 2020). Together with 14 genes associated with ear leaf width and length, 25 genes were associated with plant height. One other ortholog for a gene shown to affect phosphorus content in rice (Yamaji et al., 2017) was also included in this study.
(3) A large number of candidates derived from initially mapped QTLs for 23 important agronomic traits, identified by genome-wide association mapping studies using the recently developed population (Liu et al., 2020). For each trait, the top one or two large-effect QTLs were integrated, and genes were filtered if additional evidence (expression relevance, expression QTL associations, or ortholog information) was available; all candidates within the QTL interval were included if there was no other reliable evidence and if the interval contained less than 10 candidates. These included 243 genes associated with flowering times, 540 genes related to plant architecture traits, and another 229 and 422 genes affecting the ear and kernel-related yield traits, respectively.
(4) 270 genes from QTLs associated with dehydration rate and another seven genes potentially affecting lipid content identified by association mapping. These two studies were performed using a natural population consisting of over 500 unrelated individuals (Liu et al., 2017b).
Genes from sources (1) and (2) formed set no. 1, and two sgRNAs were designed for each gene to form the DSP. Genes from sources (2), (3), and (4) comprised set no. 2, with individual sgRNA per gene for (3) and (4) and the two sgRNAs per gene for (2) also being separately constructed; all were mixed as individual sgRNA per vector to form the SSP.
Non-Reference-Based sgRNA Design
The sgRNA oligo design criteria were fully implemented according to Lu et. al. (2017) to obtain an initial sgRNA library based on the B73 reference genome. However, due to the large genetic difference between the B73 and the transformation receptor KN5585 (a tropical line) used here, we required an additional filtering step to select those sgRNAs also suitable for KN5585. Whole-genome sequencing (∼60×) and deep mRNA sequencing (RNA-seq) on a mixture of seven tissues were used to obtain the de novo assembled contigs of KN5585, based on canonical pipelines using ABySS (Jackman et al., 2017; contig N50 = 3162) and Platanus (Kajitani et al., 2014; N50 = 565) for whole-genome sequencing and Trinity (Grabherr et al., 2011) for RNA-seq (N50 = 2167). These raw assembled contigs can be available at http://maizego.org/Resources.html (see the section “High-Throughput CRISPR/Cas9 Gene Editing”). All sgRNAs designed from the B73 genome with acceptable on-target scores were filtered by the Basic Local Alignment Search Tool (Camacho et al., 2009) against the locally assembled contigs to obtain the uniquely matched set. When the alignment between gene and sgRNA did not fully match, the sgRNAs with only one SNV or InDel were retained after replacing the given variants from KN5585. In addition, the nearly complete genomic sequences for all set no. 1 genes were PCR amplified and sequenced by the Sanger method, providing confirmation for all of their sgRNAs using this filtered method. To make this analysis friendly to a broad range of users, we developed a tool (Sun et al., 2019) with both a command-line and graphical user interface (implemented in Java) that can be easily implemented.
Vector Design, Construction, and Pooling
Three different vectors (Supplemental Figure 1) were used in this study: (1) pCPB-ZmUbi-hspCas9 came from Chuanxiao Xie (Li et al., 2017). We modified the vector construction by combining overlapping PCR and homologous recombination to obtain a single-sgRNA vector (SSV) or double-sgRNA vector in one step (Supplemental Figures 1A and 1B). In detail, pCPB-ZmUbi-hspCas9 was first linearized by HindIII. Separately, ZmU6 and the sgRNA scaffold of insertion elements were amplified through overlapping PCR with a homologous arm or sgRNA scaffold and/or 20-bp gene-specific target-attached primers. Additionally, homologous arms that match linearized pCPB-ZmUbi-hspCas9 were also added to the insertion fragment in the overlap PCR. Finally, different gene-specific insertion fragments were incorporated into pCPB-ZmUbi-hspCas9 as SSV and double-sgRNA vector. It is worth noting that the HindIII restriction enzyme recognition site was maintained in each construct so that gene-specific elements can be inserted (Li et al., 2017). pCXB052 was modified from a vector designed for genome-wide editing in rice (Lu et al., 2017) by replacing the rice promoters with the RNA polymerase II promoter of the maize ubiquitin gene (ZmUbi) and the RNA polymerase III promoter ZmU6 (Supplemental Figure 1C). pCXB053 was extended from pCPB-ZmUbi-hspCas9 through the preassembled ZmU6 and sgRNA scaffold. The difference between pCXB052 and pCXB053 was that both hspCas9 and the selection marker Basta gene (BlpR) are expressed by ZmUbi in pCXB052, and alternatively expressed by ZmUbi and enhanced Cauliflower mosaic virus 35S promoters in pCXB053. Unlike the construction approach in DSP, SSV of SSP was produced by oligo annealing and T4 ligase ligation. pCXB052 or pCXB053 was cleaved by BsaI to ligate with the sgRNA anneal products. Only the positive strains survive since the toxin ccdB gene was replaced by sgRNA. Self-ligated vectors were eliminated, which ensured that all of the clones obtained were positive and allowed for a pooled plasmid cloning. In brief, CPB-ZmUbi-hspCas9 was used for DSP, which was suitable for a single vector containing one or multiple sgRNAs. Thus, DSP was a uniform concentration (ng/μl, measured by NanoDrop2000) mixture of each Sanger-validated plasmid. The pCXB052 and pCXB053 vectors were designed for pooled CRISPR/Cas9-based knockout, since this allowed pooled ligation reaction cloning, so SSP was pooled prior to E. coli transformation.
Plasmid Pool Sequencing
The Tn5 transposase (Nanjing Vazyme Company of China, cat. no. TD501) was used to fragment mixed plasmids. For each reaction, 50 ng DNA was aliquoted with 10 μL 5× TruePrep Tagment Buffer L, 5 μL Tn5. Double-distilled water was added to 50 μL, mixed well, then incubated at 55°C for 10 min. DNA was purified with VAHTS DNA clean beads (Nanjing Vazyme Company of China, cat. no. N411-03-AA). For PCR amplification, we mixed 24 μL purified DNA, 10 μL 5× TruePrep Amplify Buffer, 5 μL PCR Primer Mix, 5 μL N5 primer, and 5 μL N7 primer, added 1 μL TruePrep Amplify Enzyme, and mixed well. The PCR program consisted of (1) 72°C for 3 min, (2) 98°C for 30 s, (3) six cycles of 98°C for 15 s, 60°C for 30 s, 72°C for 1 min, (4) 72°C for 5 min and hold at 4°C. Finally, purification was done with two rounds of VAHTS DNA clean beads (Nanjing Vazyme Company of China, cat. no. N411-03-AA), first round with 0.6× (30 μL) and second round 0.15× (7. 5μL) to collect the 300∼700 bp PCR products. The beads were eluted in 16 μL double-distilled water. The libraries that passed quality checks were subjected to the Illumina X-Ten sequencer with pair-end 150 bp.
Agrobacterium-Mediated Pooled Transformation
The plasmids were electroporated into Agrobacterium tumefaciens strain EHA105. Agrobacterium-mediated maize transformation is illustrated in Supplemental Figure 15. Maize immature embryos (IEs) of 1.5 to 1.8 mm were isolated from ears harvested 10 d after pollination into 2.0-mL tubes with 1.8 mL inoculation medium (Sidorov and Duncan, 2009) and were infected with Agrobacterium suspension (inoculation medium with 200 μM of acetosyringone and Agrobacterium cells) for 5 min, then poured onto cocultivation medium. The extra liquid was removed with pipettes. Immature embryos were placed with scutellum side up on the medium and incubated in the dark at 23°C for 48 to 72 h of cocultivation. After cocultivation, immature embryos were transferred to the resting medium and cultured for 5 to 7 d. Calluses were then transferred to the selection medium (glufosinate-ammonium 10 mg/L), incubated in the dark at 28°C for 2 weeks and transferred to fresh selection medium for another 2 weeks. Resistant calluses obtained were placed on the regeneration medium, incubated under 5000 lux at 25°C for 14 to 21 d. Regenerated shoots were transferred to rooting medium under 5000 lux at 25°C for 14 d. Leaves were sampled for PCR analysis before the plantlets were planted into greenhouse. The transformation experiments were conducted by Wimi Biotechnology.
Assigning Associated Targets to T0 Plants
The minimum number of T0 plants was determined to be ∼4 times of the number of vectors to cover most of the targets, as below simulation analysis suggested. For high-throughput detection of gene-edited plants (T0 generation), we added different barcode sequences (at least two mismatches between any two) to the ends of the universal primers (forward primer, CGTTTTGTCCCACCTTGACT; reverse primer, TTCAAGTTGATAACGGACTA) to produce amplicons, and the length of PCR amplification products was 165 bp (Supplemental Figure 4). A total of 30 forward and 96 reverse amplification primers ligated with barcodes designed to represent a maximum of 2880 lines for each batch (Supplemental Data Set 2). A forward amplification primer and 96 reverse amplification primers were used to amplify the DNA of gene-edited plants in a 96-well PCR plate. PCR products purified with DNA clean kit (ZYMO RESEARCH, cat. no. D4013) were used for library construction. DNA libraries were constructed according to the Truseq DNA low-throughput sample preparation kit (Illumina, FC-121-3001), end repair, “A” base addition, Illumina adapters ligation, and PCR enrichment followed with purification by AMPure XP beads (Supplemental Figure 4). All the DNA was extracted from seedling leaves unless otherwise specified.
The matched barcode sequences and amplified sgRNA were obtained by pair-end short-reads sequencing, so that the T0 individuals can be associated with their corresponding candidate genes, as long as contamination is avoided. To reduce the potential for contamination, we have focused on experimental design and bioinformatic analysis parameters affecting the reliability. Through mixing several lines with individually transformed sgRNA and negative controls (wild-type tissue, water, and empty wells), iterative sequencing with various coverage was performed. Four parameters were considered (Supplemental Figure 5A), including supported reads (count_cutoff from 5 to 200), relative ratio of supported reads at given well (ratio_cutoff, from 0.01 to 0.2), inflection point of relative amount (fold change between ratios) between sorted targets (the largest fold change of N+1th target compared to the Nth target for all targets that meet the requirements of count_cutoff and ratio_cutoff, named as peakFC), and the fold enrichment of target among the whole 96-plates, relative to mean (measured as contamination, the targets would be iteratively removed with cutoff decreasing from 5 decreases to 1.5 with a step of 0.5).
Adequate sequencing coverage is essential for eliminating background noise. While the false-negative rates were usually low, the false-positive rate is sensitive to floating count and ratio cut-offs and highly correlated to total effective discovery number (Supplemental Figures 5B to 5E). That is, a strict cut-off would lead to lower false positives, but at the cost of reducing total effective assignments. By sequencing multiple biological and technical replicates, a stricter cut-off is possible, increasing reproducibility. Taken together, targets that passed the relatively strict cut-offs (count_cutoff = 100, ratio_cutoff = 10%, targets ranked above the peakFC, contamination_cutoff = 2× mean coverage of each individual) and were identified in at least two repeats were used to ensure high-confidence assignments. However, all of the remaining sgRNAs identified in only one experiment were also incorporated in mutated sequence detection, even though very few were validated by mutants.
Simulation of Target Coverage as a Function of the Number of T0 Individuals
Considering the transformation and planting limitation, it is important to balance the plant pool size and gene/target coverage of each pooled transformation assay. To decide how many genes/vectors (Vn) should be mixed in a pool, we performed a simulation, with Vn from 1 to 200 and the number of T0 individuals (Pn) from 1 to 10 times Vn. Fifty replicates of the primary vector pool were created as follows. Vectors were randomly selected from the amplified vector pool without replacement, to obtain Vns. Finally, the coverage was calculated as the ratio to Vn. The simulation for a given vector pool and plant library was repeated 100 times, and three values (mean, minimum, and standard value) were considered to select the primary vector mixture size and the number of plants needed.
From the simulation analysis and the observed cases of coverage of sgRNAs along various T0 lines, four times the number of T0 plants (relative to gene/vector number) were required to cover most of the candidates, comparable with observed results. Given a 50-vector pool as an example, 98.7% of genes on average (with a min of 94%) can be covered by 200 (4×) T0 lines (Supplemental Figure 6), and the coverage was better for a larger number of vector pools. However, over half of the genes (or vectors) were present in fewer than three plants, and 30% were represented by a single individual. This distribution represented a risk in further experiments (including the identification of effective mutant alleles, independent cross validations, or even collection of sufficient seeds for next generation); 10 times the number of T0 plants would then be needed to represent more than 85% of genes by at least three lines.
Identification of Mutated Alleles by Sanger Sequencing
Sanger sequencing was applied for all amplicons to obtain “.ab1” files, and the R package sangerseqR (Hill et al., 2014) was used for base calls and plotting chromatograms. By using the Poly Peak Parser, this package can separate ambiguous base calls into two sequences. A ratio = 0.2 was set for separating signal and noise base calls, and the 20 bp at the beginning and end of the sequence were trimmed when generating chromatogram plots. The obtained primary and secondary sequences were considered as two haplotypes, which are identical for homozygous mutations. Further analyses were the same for homozygous or heterozygous mutations. The primary and secondary sequences together with the wild-type genomic and sgRNA sequences were used as input to multiple sequence alignment by Clustal programs (Larkin et al., 2007) to call specific variants. It is important to note that both the forward and reverse amplicons help identify exact alleles or at least to clarify the mutated position/intervals. However, for those lines containing more than two mutated alleles, this method will not uncover separate alleles.
Identification of Mutated Alleles by MassARRAY
We used MassARRAY technology to genotype known variants for multiple loci in large populations. An introduction to MassARRAY, laboratory protocol, and analysis is available at http://agenabio.com/products/massarray-system. Based on the conventional MassARRAY process, we applied a sequential primer combination strategy (Supplemental Figures 7 and 8) to detect if given nucleotides are altered, resulting in an opportunity to infer the likely mutants by integrating all the sequential outcomes. All the experiments in this study were performed by Agena Bioscience in Beijing. Based on the design of a primer covering the predicted nuclease cleavage region (3 to 6 bp upstream of the NGG PAM sequence), this method is preferable to the determination of whether individuals of interest were mutated at given genes or to the identification of known variants at the T1 or later generations in a large number of individuals. A full comparison of the advantages and disadvantages of Sanger sequencing, the MassARRAY method, and capture sequencing are described in Supplemental Table 2.
Identification of Mutated Alleles by Capture Sequencing
Targeted capture was realized by GenoPlexs technology, which captures multiple target regions using a set of primer pairs and a single polymerase chain reaction. All the capture primers were designed by MOLBREEDING. After removing genes with difficulties in primer design and primers with low efficiency or nonspecificity, we retained a total of 106 genes with 113 primer pairs (Supplemental Data Set 3) for further analysis. Deep pair-end (PE) sequencing (>500×) on the captured products was performed on an Illumina HiSeq 3000. All reads were trimmed by Trimmomatic (Bolger et al., 2014) with the following parameters: LEADING:5 TRAILING:5 SLIDINGWINDOW:3:20 MINLEN:50, and only clean PE reads were used in the next analysis.
As all the T0 individuals had been assigned to corresponding targets, lines with different targets can be mixed in capture sequencing to reduce library construction cost. By applying modeling with three wild-type line repeats and varying numbers (5∼50) of mixed individuals, we found a mix of 20∼25 lines would be best, with a 0.3% ratio of background mutant error, presumably because of aerosol contamination and PCR or sequencing errors.
The CRISPResso2 software (Clement et al., 2019) was applied for the identification of mutated alleles and estimation of their frequencies. Only the mutations that overlapped with the 20-bp window before the NGG PAM were considered unless the subsequent analysis detected likely alleles caused by HDR, in which case flanking variants were also considered. The abridged sequences within the 20-bp window were merged when identical. The alleles supported by less than three reads and those present in wild samples (including three technical repeats) were discarded in further analysis, and allele-specific frequencies were re-estimated when there was more than one allele. A variant-calling pipeline was also integrated in allele identification: the clean PE reads were first mapped to pseudogenome (derived from replacing specific variants to B73 genome) by bwa-mem (Li, 2013), followed by SNP and InDel calling using the mpileup command from samtools (Li et al., 2009) at all target regions.
To avoid assigning identical mutants to different alleles as a result of ambiguous alignments, entire mutated sequences were used to determine whether the alleles called were consistent between different methods. All the different alignments from the identical alleles were assumed to be the one with overlap (or close) to the predicted nuclease cleavage site, as CRISPResso2 (Clement et al., 2019) suggested.
Testing the Predictability of Edited Outcomes
All of the alleles with precise variant sequences from both SSP and DSP pools and both Sanger and capture sequencing methods were merged as two data sets, one containing all of the mutants occurring at individual sgRNA, the other containing large fragment mutants (deletion, insertion, and reversion) between pair sgRNAs. The mutant type (DEL, INS, or SNP), position (relative to predicted nuclease cleavage site), and size (for DEL and INS) were considered to be characteristic of a variant, while the 20-bp sgRNA nucleotides and the PAM sequences, as well as the target gene’s expression quantification (data from Chen et al. [2014]), number of tissues with expression of fragments per kilobase of exon model per million reads mapped > 0.5) and expression variability along developmental period (measured by coefficient of variation) were all regarded as predictive variables (Supplemental Figure 10A). The Random Forest algorithm, which is nonparametric, interpretable, and compatible with many types of data with high prediction accuracy, was applied in prediction tests from sgRNA sequences and target expression variables. The out-of-bag error and mean of squared residuals were used to evaluate the predictability for classification (mutant type) and the regression variables (mutant position and size), respectively. The Gini decreases (MeanDecreaseGini) and node purity increase (IncNodePurity) values for each variable over all trees were used to evaluate the variable importance for classification (mutant type) and the regression variables (mutant position and size), respectively.
The prediction algorithm FORECasT (favored outcomes of repair events at Cas9 targets; Allen et al., 2018), fine-tuned using over 109 mutational outcomes from over 40,000 human sgRNAs, was used in predicting likely repair outcomes by flanking DNA sequence. First, the effect of the lengths of flanking sequences (10, 20, 50, 100) on allele prediction was examined. While they generally produced highly replicable results, a longer flanking region led to a higher number of predicted alleles with rare frequency. Nevertheless, there was no effect when the flanking region was greater than 50 bp, as predictions with 50 bp and 100 bp being identical. Thus, all the results from this set were used in further analysis. The entire mutated sequence incorporated with variants together with corresponding predicted frequencies were used to compare with those real observed alleles.
Discovery of Alleles Likely Derived from HDR
Those mutated haplotypes with concurrent InDels at the sgRNA region and at least two SNPs within flanking sequences were considered a possible consequence of HDR. These mutated sequences were then compared by the Basic Local Alignment Search Tool to all the de novo assembled contigs to search for a likely template source.
Identification of Expression Compensation of ZmBAM1d Mutant Lines by RNA Sequencing
ZmBAM1d (Zm00001d028317) was edited with two sgRNAs targeting the first exon. RNA sequencing on whole kernel (20 d after pollination) was performed for self-crossed T3 edited lines with homozygous fragment deletion and wild-type lines, both with three replicates. Raw reads were first trimmed with Trimmomatic (Bolger et al., 2014). All remaining paired-end clean reads were mapped to the B73_V4 reference genome (Jiao et al., 2017) using Tophat2 (Kim et al., 2013). The Cuffquant and Cuffdiff (Trapnell et al., 2013) commands from Cufflinks (Trapnell et al., 2010; Roberts et al., 2011) were used to estimate RNA abundance and to test for differential expression, respectively. The geometric method was used to normalize the fragments per kilobase of exon model per million reads mapped across all libraries (Anders and Huber, 2010) during differential expression analysis.
Off-Target Analysis
A total of 20 T1 blocks with dramatic phenotypic changes were selected to measure the off-target effect, with at least four individual T1 lines from the same T0 background mixed to represent each sample. Genomic DNA was isolated from mature leaves. DNA extraction and library construction were the same as above, with an additional hybridization process with the Roche/NimbleGen SeqCap EZ library, which was specifically designed to capture the exon sequences of maize by high-density biotinylated long oligonucleotide probes. The BGISEQ-500 platform was used in paired-end 150 bp short-reads sequencing.
All the clean reads trimmed by Trimmomatic (Bolger et al., 2014) were aligned to the B73_V4 reference genome by BWA-mem (Li, 2013). Variants were called by GATK HaplotypeCaller (Poplin et al., 2018) with Genomic Variant Call Format mode. Only InDels supported with at least three reads for each sample were conserved. Those variants were discarded in further analyses if they (1) also were called by wild-type lines against the B73 reference genome (background genetic variations), or (2) “ALT” alleles were simultaneously present in over three lines (common variants). The remaining InDels located within all potential targets were considered as on-targets. One sample was abandoned since no likely on-target loci were found. The remaining 19 samples targeted a total of 25 genes. The Cas-OFFinder (Bae et al., 2014) was used to predict the corresponding off-target loci, with at most five mismatches and NGG PAM. Those InDels located within these possible off-target regions were regarded as likely off-targeting events.
Phenotyping
All the T0 individuals were self-crossed if conditions allowed or backcrossed to wild lines (KN5585) if self-crossing was not possible due to phenotypes affecting reproductive structures (of which information was all recorded). Generally, at least two independent events were planted if available. For the DSP gene set, all the T0 plants were first inspected for mutated alleles (DNA from seedling leaf), and those events with clearly edited sequences resulting in likely nonfunctional alleles were planted with expanded T1 or greater populations. For the SSP gene set, all the T0 events with seed numbers larger than 10 (including lines that failed target assignment) were planted for phenotyping, and the lines with observed agronomic trait variance were genotyped. We planted 17 genotyped individuals per cell for phenotyping during the T1 generation. Wild-type controls were planted every 4 to 30 rows based on specific designs, variation in the number of total events, and space limitations. Phenotypic differences relative to wild type and segregating independently within T1 lines that were from the same T0 event were recorded as heritable phenotypic changes. Multiple locations (from northeast temperate to southwest and south tropical zone, including Gongzhuling city, Jilin province, 43°30′N 124°49′E; Gasa town, Xishuangbanna dai autonomous prefecture, Yunnan province, 21°57′N 100°45′E; and Foluo town, Sanya City, Hainan Province, 18°34′N 108°43′E) were used to evaluate the environmental effect for DSP; however, only the Beijing location (in summer of 2018) was used in the large-scale measurement of the T1 performance for SSP.
Genetic Materials Module
In addition to the general considerations listed above, the examples used in interpreting genotype-phenotype links are described in detail here. Mutants of zmtps14.1 were from DSP (two sgRNAs are simultaneously designed), whose phenotypic change was supported by large fragment deletion F2 populations at Hainan (south China; 61 mutant lines versus 173 wild lines; Figure 4B; Supplemental Figure 12B). The zmsbp22 was supported by six independent T1 populations (derived from DSP, 52 positive/mutant lines versus 20 negative/wild lines) at Yunan (southwest China; Figure 4C; Supplemental Figure 12B), and two mutant alleles from SSP (only one sgRNA is used) along with considering all the other lines as “control” (10 target gene mutant lines compared to all the other 470 lines with various mutant genes; Supplemental Figure 12D) were compared for double confirmation. The example in the aminotransferase-like gene GRMZM2G331652 was supported by data from both T1 (62 positive versus 17 negative lines) and T2 data at two locations (39 mutants versus 30 wild lines at Hainan; 39 mutants versus 45 lines at Jilin; Supplemental Figures 13B to 13D). For the zmbam1d, self-crossed T3 lines with a large fragment deletion (from two sgRNAs) were used to measure kernel weight (Figures 4D and 4E) at Yunnan (five mutants versus 13 wild ears) and Wuhan (central China; 39 mutants versus 10 wild ears). Detailed phenotypes for these examples are provided in Supplemental Data Set 4 .
For those “unexpected” mutant lines shown in Supplemental Figure 14, at least two individuals showing mutant phenotypes and separated within T1 populations (from same T0) or the whole T1 population displayed significant differences relative to wild types are considered as heritable (but not environmental) phenotypic changes. For T1 or advanced populations, we did not evaluate for the presence of a transgene, but instead, we detect the target alleles for all the phenotyped lines using mature leaves as source for DNA.
The vectors used in present study can be requested from J.X. (xjt@wimibio.com). All the information of the mutants are available at the official website of WIMI Biotechnology (http://www.wimibio.com/tbtk.asp), which will be continuously updated and the seeds can be requested with the standard MTA (http://www.wimibio.com/e.doc) and specified charge.
Software/Custom Scripts
The CRISPR-Local for high-throughput designing sgRNAs for nonreference lines can be obtained from: https://github.com/sunjiamin0824/CRISPR-Local.git. And the script to obtain reads that matched both the barcodes and pooled sgRNAs from trimmed fastq files is available at https://github.com/heroalone/crispr_pool.git.
Accession Numbers
Raw whole-genome sequencing and RNA sequencing reads of the transformation receptor (KN5585), and raw reads of TRC-seq for 60 batches have been deposited in the Genome Sequence Archive (Wang et al., 2017b) of BIG Data Center (BIG Data Center Members, 2017) under the following accession number: CRA001955 (https://bigd.big.ac.cn/gsa/browse/CRA001955). Individual fastq files can be downloaded under the “Run Accession” links. Assembled contigs can be downloaded at http://maizego.org/Resources.html (“High-Throughput CRISPR/Cas9 Gene Editing” section).
SUPPLEMENTAL DATA
Supplemental Figure 1. Structure of all constructs.
Supplemental Figure 2. The mutation rate for the ZmPLA1 gene of the primary vector, pCPB-ZmUbi-hspCas9.
Supplemental Figure 3. Different strategies and relevant data generated of SSP and DSP.
Supplemental Figure 4. Barcode-based NGS in target identification.
Supplemental Figure 5. Selection of sequence cut-off parameters to ensure the reliability of target determination.
Supplemental Figure 6. Simulation and analysis of the sgRNA coverage along various T0 plants.
Supplemental Figure 7. Use of the MassARRAY method in mutant sequence identification.
Supplemental Figure 8. Mutant sequences inferred by the MassARRAY method.
Supplemental Figure 9. Mutation patterns and predictability of deletion occurring between pair sgRNAs.
Supplemental Figure 10. Prediction of mutations within sgRNA sequences and target expression variables using Random Forest.
Supplemental Figure 11. Identification of mutants caused by HDR.
Supplemental Figure 12. Identification of genes affecting maize flowering time.
Supplemental Figure 13. Identification of phenotypic changes in mutants inconsistent with results of association mapping.
Supplemental Figure 14. Identification of a representative set of unexpected phenotypic variations.
Supplemental Figure 15. Agrobacterium-mediated transformation using maize immature embryos.
Supplemental Table 1. Transformation frequencies for different vectors.
Supplemental Table 2. Comparison of the advantages and shortcomings of different methods in mutant sequence identification.
Supplemental Data Set 1. List of mutant alleles with their variant sequences.
Supplemental Data Set 2. Barcode sequences used in target determination.
Supplemental Data Set 3. Primer pairs used in capture-sequencing.
Supplemental Data Set 4. Detailed phenotypes for the referred biological examples.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We would like to thank Chuanxiao Xie from the Institute of Crop Science of the Chinese Academy of Agricultural Sciences for providing the basic vector, Jia’nan Zhang from the MOLBREEDING company (Shijiazhuang, China) for designing the capture primers, and Gehua Liu from GENOSTAR (Beijing, China) and Xin He from Agena Bioscience (Beijing, China) for supporting the experiments by using MassARRAY. We thank other colleagues from WIMI Biotechnology for helping with bench work. This research was supported by the National Transgenic Major Project of China (2018ZX08010-04B, 2019ZX08010003-002-013), the National Natural Science Foundation of China (31525017, 31961133002, 31901553), the National Key Research and Development Program of China (2016YFD0101003), the Postdoctoral Talent Innovation Program of China (BX201700092), and Fundamental Research Funds for the Central Universities. J.X., B.H., L.H., X.Y., and Y.Q. are employees of WIMI Biotechnology Co., Ltd. Y.Lu is an employee of Biogle Genome Editing Center. H.-J.L., J.X., and Jianbing Y. have filed a provisional patent related to the improved MassARRAY method in edited allele identification.
AUTHOR CONTRIBUTIONS
Jianbing Y. designed and supervised this study. L.J., J.X., Mi.Z., and X.S. constructed the plasmids. J.X., L.H., and X.Y. performed the transformation and positive transgenic line filtering. Y.Lu, J.S., and H.-J.L. designed the line-specific sgRNAs. Q.Z., Y.P., and Jiali Y. performed all library construction and NGS sequencing. H.-J.L. performed most of the bioinformatics analyses. W.W. performed the off-target analysis. B.H., F.G., Y.D., and Z.L. were responsible for planting T0-positive transgenic lines in the greenhouse and field. Ma.Z., M.J., X.L., M.B., W.S., Y. Liu, J.Z., W.L., and H.-J.L. performed the field trail and phenotyping. Q.Z. and H.-J.L. performed genotyping by Capture sequencing. M.J., Jiali Y., and H.C. performed genotyping by Sanger sequencing. J.Liu conducted zmbam1d analysis. Y.Q. assisted the management of the whole project. H.-J.L., L.J., D.J., A.R.F., and Jianbing Y. made in-depth discussions and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Allen F., et al. (2018). Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 37: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W.(2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayar A., Wehrkamp-Richter S., Laffaire J.B., Le Goff S., Levy J., Chaignon S., Salmi H., Lepicard A., Sallaud C., Gallego M.E., White C.I., Paul W.(2013). Gene targeting in maize by somatic ectopic recombination. Plant Biotechnol. J. 11: 305–314. [DOI] [PubMed] [Google Scholar]
- Bae S., Park J., Kim J.S.(2014). Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M., et al. (2019). Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soybean. Plant Biotechnol. J. 18: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIG Data Center Members (2017). The BIG Data Center: From deposition to integration to translation. Nucleic Acids Res. 45 (D1): D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B.(2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L., et al. (2016). Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 14: 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E.S., et al. (2009). The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.(2009). BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A.M., Henser-Brownhill T., Monserrat J., Poetsch A.R., Luscombe N.M., Scaffidi P.(2019). Target-specific precision of CRISPR-mediated genome editing. Mol. Cell 73: 699–713.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zeng B., Zhang M., Xie S., Wang G., Hauck A., Lai J.(2014). Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C.(2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70: 667–697. [DOI] [PubMed] [Google Scholar]
- Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E., Pinello L.(2019). CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37: 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles N.D., McMullen M.D., Balint-Kurti P.J., Pratt R.C., Holland J.B.(2010). Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics 184: 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F.(2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite M., Park W., Xu Z., Yan X., Maurais S.C., Dooner H.K.(2002). Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.A., Ong B.(2017). The MassARRAY® system for targeted SNP genotyping. Methods Mol. Biol. 1492: 77–94. [DOI] [PubMed] [Google Scholar]
- Fang H., et al. (2020). Genetic basis of kernel nutritional traits during maize domestication and improvement. Plant J. 101: 278–292. [DOI] [PubMed] [Google Scholar]
- Fauser F., Roth N., Pacher M., Ilg G., Sánchez-Fernández R., Biesgen C., Puchta H.(2012). In planta gene targeting. Proc. Natl. Acad. Sci. USA 109: 7535–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Yan J.(2019). De novo domestication: An alternative route toward new crops for the future. Mol. Plant 12: 615–631. [DOI] [PubMed] [Google Scholar]
- Gil-Humanes J., Wang Y., Liang Z., Shan Q., Ozuna C.V., Sánchez-León S., Baltes N.J., Starker C., Barro F., Gao C., Voytas D.F.(2017). High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., et al. (2018). Stepwise cis-regulatory changes in ZCN8 contribute to maize flowering-time adaptation. Curr. Biol. 28: 3005–3015.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.T., Demarest B.L., Bisgrove B.W., Su Y.C., Smith M., Yost H.J.(2014). Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev. Dyn. 243: 1632–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K., Zhang J., Botella J.R., Ma C., Kong F., Liu B., Zhu J.K.(2019). Perspectives on the application of genome-editing technologies in crop breeding. Mol. Plant 12: 1047–1059. [DOI] [PubMed] [Google Scholar]
- Jackman S.D., Vandervalk B.P., Mohamadi H., Chu J., Yeo S., Hammond S.A., Jahesh G., Khan H., Coombe L., Warren R.L., Birol I.(2017). ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22: 561–570. [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2017). Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T., Lowe K., Hoerster G., Anand A., Wu E., Wang N., Arling M., Lenderts B., Gordon-Kamm W.(2019). Maize transformation using the morphogenic genes Baby Boom and Wuschel2. Methods Mol. Biol. 1864: 81–93. [DOI] [PubMed] [Google Scholar]
- Kajitani R., et al. (2014). Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.(2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lazakis C.M., Coneva V., Colasanti J.(2011). ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J. Exp. Bot. 62: 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Liu C., Qi X., Wu Y., Fei X., Mao L., Cheng B., Li X., Xie C. (2017). RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol. J. 15: 1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997v1.
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., et al. (2019a). Whole genome sequencing reveals rare off-target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR/Cas9-edited cotton plants. Plant Biotechnol. J. 17: 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang Y., Chen K.L., Shan Q.W., Wang Y.P., Liang Z., Gao C.X.(2013). [CRISPR/Cas: A novel way of RNA-guided genome editing]. Yi Chuan 35: 1265–1273. [DOI] [PubMed] [Google Scholar]
- Li S., Li J., He Y., Xu M., Zhang J., Du W., Zhao Y., Xia L.(2019b). Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat. Biotechnol. 37: 445–450. [DOI] [PubMed] [Google Scholar]
- Liang Y., et al. (2019). ZmMADS69 functions as a flowering activator through the ZmRap2.7-ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol. 221: 2335–2347. [DOI] [PubMed] [Google Scholar]
- Lieberman-Lazarovich M., Levy A.A.(2011). Homologous recombination in plants: An antireview. Methods Mol. Biol. 701: 51–65. [DOI] [PubMed] [Google Scholar]
- Liu C., et al. (2017a). A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 10: 520–522. [DOI] [PubMed] [Google Scholar]
- Liu H., Luo X., Niu L., Xiao Y., Chen L., Liu J., Wang X., Jin M., Li W., Zhang Q., Yan J.(2017b). Distant eQTLs and non-coding sequences play critical roles in regulating gene expression and quantitative trait variation in maize. Mol. Plant 10: 414–426. [DOI] [PubMed] [Google Scholar]
- Liu H.J., et al. (2020). CUBIC: An atlas of genetic architecture promises directed maize improvement. Genome Biol. 21: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.J., Yan J.(2019). Crop genome-wide association study: A harvest of biological relevance. Plant J. 97: 8–18. [DOI] [PubMed] [Google Scholar]
- Liu Z., et al. (2016). Expanding maize genetic resources with predomestication alleles: Maize-teosinte introgression populations. Plant Genome 9. [DOI] [PubMed] [Google Scholar]
- Lowe K., La Rota M., Hoerster G., Hastings C., Wang N., Chamberlin M., Wu E., Jones T., Gordon-Kamm W.(2018). Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 54: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28: 1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., et al. (2018). Gene-indexed mutations in maize. Mol. Plant 11: 496–504. [DOI] [PubMed] [Google Scholar]
- Lu Y., Ye X., Guo R., Huang J., Wang W., Tang J., Tan L., Zhu J.K., Chu C., Qian Y.(2017). Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 10: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.(2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B.P., Liu H., Vollbrecht E., Senior L., Rabinowicz P.D., Roh D., Pan X., Stein L., Freeling M., Alexander D., Martienssen R.(2003). Maize-targeted mutagenesis: A knockout resource for maize. Proc. Natl. Acad. Sci. USA 100: 11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. [DOI] [PubMed] [Google Scholar]
- Meng X., Muszynski M.G., Danilevskaya O.N.(2011). The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23: 942–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Yu H., Zhang Y., Zhuang F., Song X., Gao S., Gao C., Li J.(2017). Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant 10: 1238–1241. [DOI] [PubMed] [Google Scholar]
- Mitter N., Worrall E.A., Robinson K.E., Li P., Jain R.G., Taochy C., Fletcher S.J., Carroll B.J., Lu G.Q., Xu Z.P.(2017). Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 3: 16207. [DOI] [PubMed] [Google Scholar]
- Nekrasov V., Staskawicz B., Weigel D., Jones J.D., Kamoun S.(2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 691–693. [DOI] [PubMed] [Google Scholar]
- Peng J., Zhou Y., Zhu S., Wei W.(2015). High-throughput screens in mammalian cells using the CRISPR-Cas9 system. FEBS J. 282: 2089–2096. [DOI] [PubMed] [Google Scholar]
- Peng Y., et al. (2019). Chromatin interaction maps reveal genetic regulation for quantitative traits in maize. Nat. Commun. 10: 2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R., et al. (2018). Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178. [Google Scholar]
- Puchta H.(1999). Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics 152: 1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Dong L., Liu C., Mao L., Liu F., Zhang X., Cheng B., Xie C.(2018). Systematic identification of endogenous RNA polymerase III promoters for efficient RNA guide-based genome editing technologies in maize. Crop J. 6: 314–320. [Google Scholar]
- Ray D.K., Mueller N.D., West P.C., Foley J.A.(2013). Yield trends are insufficient to double global crop production by 2050. PLoS One 8: e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L.(2011). Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D., Molinier J., Fritsch O., Hohn B.(2005). The dual nature of homologous recombination in plants. Trends Genet. 21: 172–181. [DOI] [PubMed] [Google Scholar]
- Settles A.M., et al. (2007). Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Zhang F.(2015). High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., Zhang K., Liu J., Xi J.J., Qiu J.L., Gao C.(2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31: 686–688. [DOI] [PubMed] [Google Scholar]
- Shou J., Li J., Liu Y., Wu Q.(2018). Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol. Cell 71: 498–509.e4. [DOI] [PubMed] [Google Scholar]
- Sidorov V., Duncan D.(2009). Agrobacterium-mediated maize transformation: Immature embryos versus callus. Methods Mol. Biol. 526: 47–58. [DOI] [PubMed] [Google Scholar]
- Spampinato C.P.(2017). Protecting DNA from errors and damage: An overview of DNA repair mechanisms in plants compared to mammals. Cell. Mol. Life Sci. 74: 1693–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Liu H., Liu J., Cheng S., Peng Y., Zhang Q., Yan J., Liu H.J., Chen L.L.(2019). CRISPR-Local: A local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics 35: 2501–2503. [DOI] [PubMed] [Google Scholar]
- Tang X., et al. (2018). A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 19: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L.(2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.(2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkuijl S.A., Rots M.G.(2019). The influence of eukaryotic chromatin state on CRISPR-Cas9 editing efficiencies. Curr. Opin. Biotechnol. 55: 68–73. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., et al. (2010). Genome-wide distribution of transposed dissociation elements in maize. Plant Cell 22: 1667–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., Feil R., Lunn J.E., Stitt M., Schmid M.(2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707. [DOI] [PubMed] [Google Scholar]
- Wang M., Lu Y., Botella J.R., Mao Y., Hua K., Zhu J.K.(2017a). Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol. Plant 10: 1007–1010. [DOI] [PubMed] [Google Scholar]
- Wang N., Long T., Yao W., Xiong L., Zhang Q., Wu C.(2013). Mutant resources for the functional analysis of the rice genome. Mol. Plant 6: 596–604. [DOI] [PubMed] [Google Scholar]
- Wang Y., et al. (2017b). GSA: Genome sequence archive<sup/>. Genomics Proteomics Bioinformatics 15: 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Liu H., Wu L., Warburton M., Yan J.(2017). Genome-wide association studies in maize: Praise and stargaze. Mol. Plant 10: 359–374. [DOI] [PubMed] [Google Scholar]
- Yamaji N., Takemoto Y., Miyaji T., Mitani-Ueno N., Yoshida K.T., Ma J.F.(2017). Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 541: 92–95. [DOI] [PubMed] [Google Scholar]
- Yang N., et al. (2019). Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet. 51: 1052–1059. [DOI] [PubMed] [Google Scholar]
- Yin K., Gao C., Qiu J.L.(2017). Progress and prospects in plant genome editing. Nat. Plants 3: 17107. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Malzahn A.A., Sretenovic S., Qi Y.(2019). The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5: 778–794. [DOI] [PubMed] [Google Scholar]
- Zhu J., Song N., Sun S., Yang W., Zhao H., Song W., Lai J.(2016). Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J. Genet. Genomics 43: 25–36. [DOI] [PubMed] [Google Scholar]