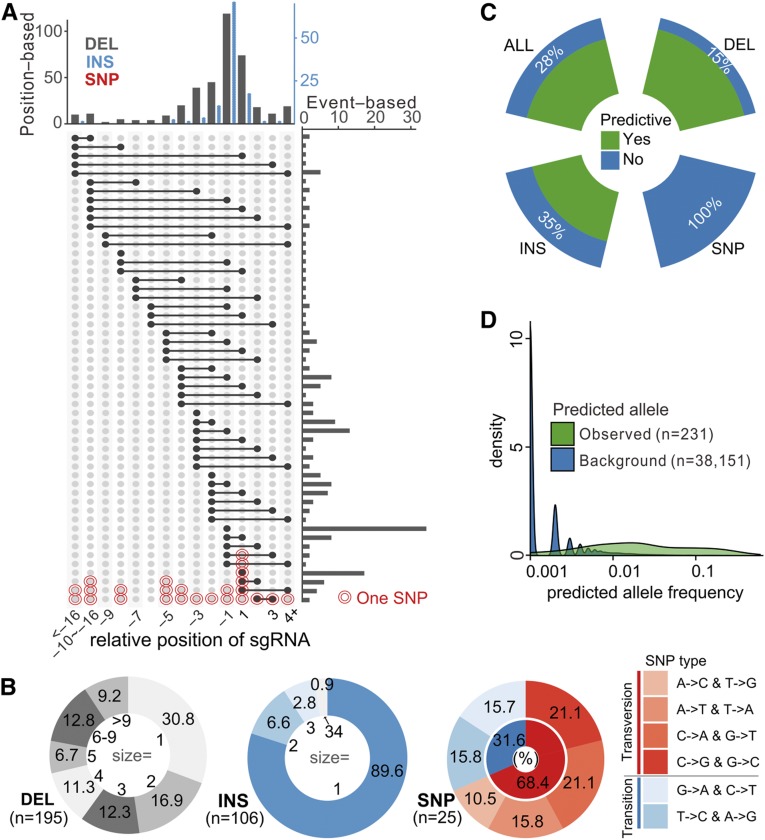
Figure 3.
Mutation Pattern and Predictability of Variants Generated by Genome Editing.
(A) Allele size and position distribution based on all individual events. Position-based, distribution along relative position on sgRNA; event-based, distribution of individual events. The position along sgRNA (x axis) is relative to predicted nuclease cleavage site, while +1 and −1 indicate the nucleotides 3 to 4 bp upstream of the PAM. INS, Insertion; DEL, deletion.
(B) Distribution of mutant outcome sizes (in bp) and diversity for different mutant classes.
(C) Ratio of real observed alleles that are being predicted by only flanking sequences, classified by mutant types (DEL, INS, SNP). ALL corresponds to all mutant types added.
(D) Algorithm-mediated prediction of mutant outcomes based on flanking sequences. The set of alleles observed in real cases display significantly higher predicted frequency compared with all predicted outcomes (background).