Fig. 4 |. Roles of the Brca1 complex in protecting regressed replication forks and resolving r-loops.
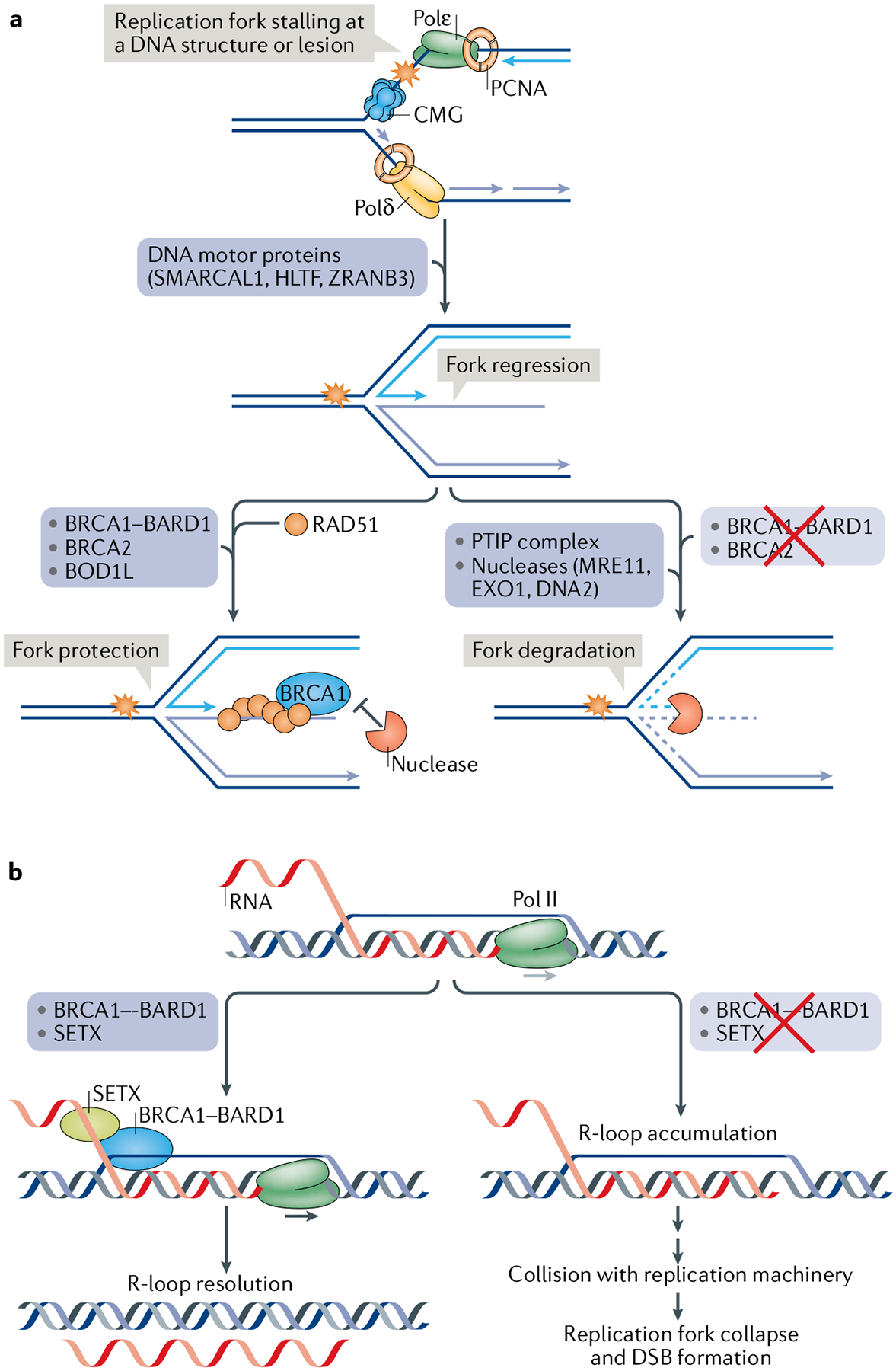
a | During DNA replication, when the DNA polymerase ensemble comprising DNA polymerase-ε (Polε), PCNA and the CDC45-MCM-GINS (CMG) helicase complex on the leading strand and Polδ-PCNA on the lagging strand is impeded by a DNA structure or lesion, one of the nucleic acid motor proteins SMARCAL1, HLTF or ZRANB3 can catalyse the reversal of the replication fork, a process often referred to as ‘replication fork regression’, to generate a four-armed DNA structure that harbours a free DNA end. Formation of the regressed fork allows a replicative mechanism of lesion bypass15,227. However, the free DNA end is prone to attack by MRE11 and other nucleases in conjunction with the PTIP complex148, unless RAD51 is deposited on the regressed fork. The stability of the regressed fork is also dependent on breast cancer type 1 susceptibility protein (BRCA1)-BRCA1-associated RING domain 1 (BARD1), BRCA2 and biorientation of chromosomes in cell division protein 1-like 1 (BOD1L), which are postulated to stabilize the RAD51-DNA interaction to prevent nucleolytic attrition139,140,146. Other pathways of replication fork repair and restart have been identified, and readers are referred to several comprehensive reviews on this topic227–229. b | Stalling of RNA polymerase II (Pol II) or delay in mRNA processing can lead to accumulation of R-loops (RNA-DNA hybrids). An R-loop can stall the DNA replication machinery, thereby causing fork collapse and the formation of a DNA double-strand break (DSB). BRCA1-BARD1 and the putative RNA-DNA helicase senataxin (SETX) facilitate R-loop resolution155.
