Fig.2. AIP1A was degraded by the Smurf1-mediated proteolytic pathway in response to TNF.
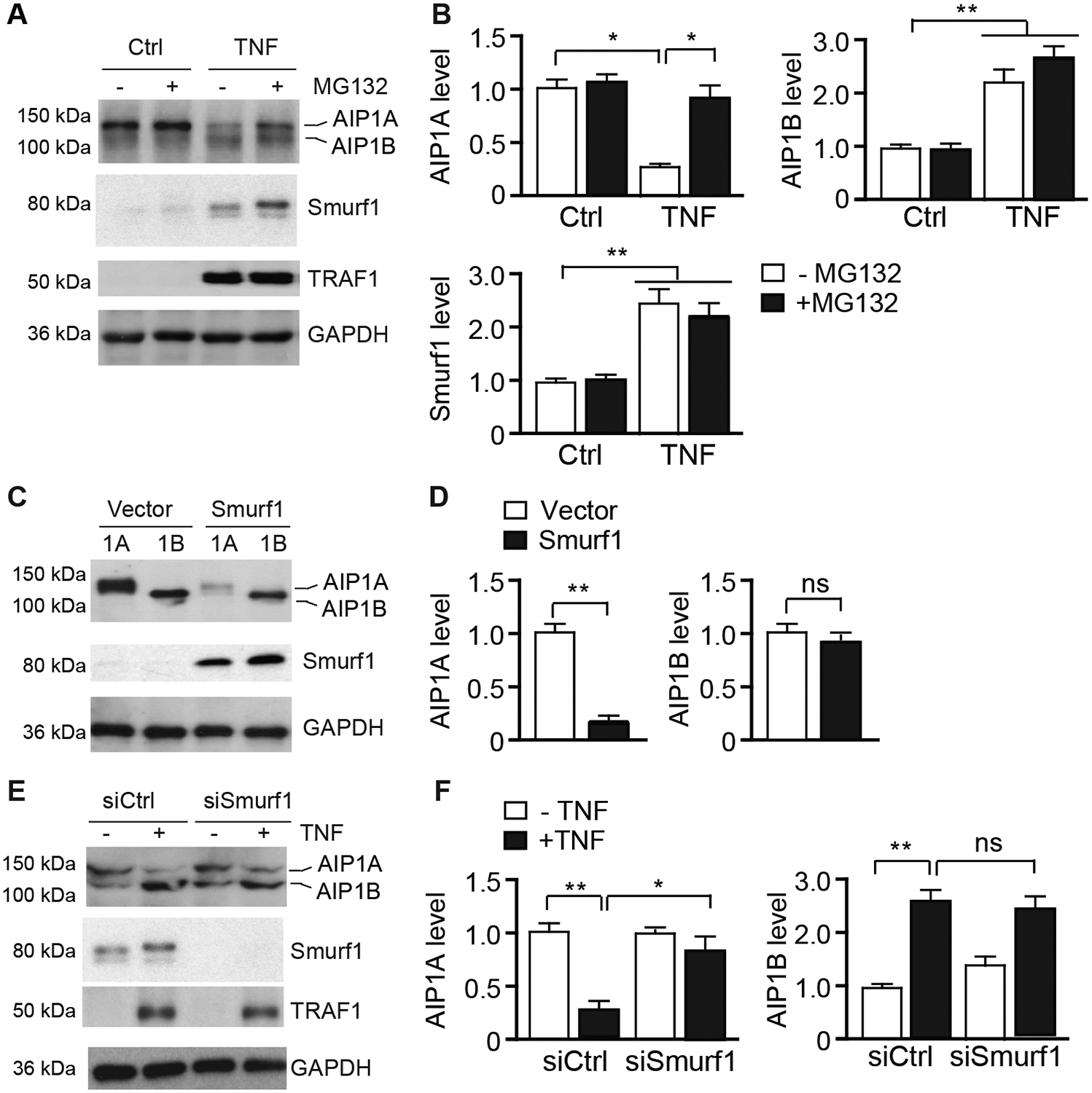
A-B. HUVECs were untreated or treated with TNF (10 ng/ml) for 16 h followed by incubation in the presence or absence of MG132 (20 μM) for 8 h. AIP1, Smurf1, and TNF-inducible TRAF1 proteins were detected by western blotting respective antibodies (A). Protein bands were quantified by densitometry and fold changes are presented by taking untreated group as 1.0 (B). n=3. C-D. HUVECs were transfected with an expression plasmid for AIP1A or AIP1B in the presence or absence of Smurf1 expression plasmid for 48 h. AIP1 expression was determined by western blotting with anit-pan-AIP1 (C). AIP1 protein bands were quantified by densitometry and fold changes are presented by taking vector group as 1.0 (D). n=3. E-F. HUVECs were transfected with control or Smurf1 siRNA for 48 h. Cells were then untreated or treated with TNF (10 ng/ml) for 8 h. AIP1, Smurf1, and TRAF1 proteins were detected by western blotting (E). AIP1 protein bands were quantified by densitometry and fold changes are presented by taking untreated group as 1.0 (F). All data are presented as the mean ± SEM. * P<0.05, ** P<0.01, one-way ANOVA followed by Bonferroni’s post-hoc test (B) or unpaired, two tailed t-test (D,F).
