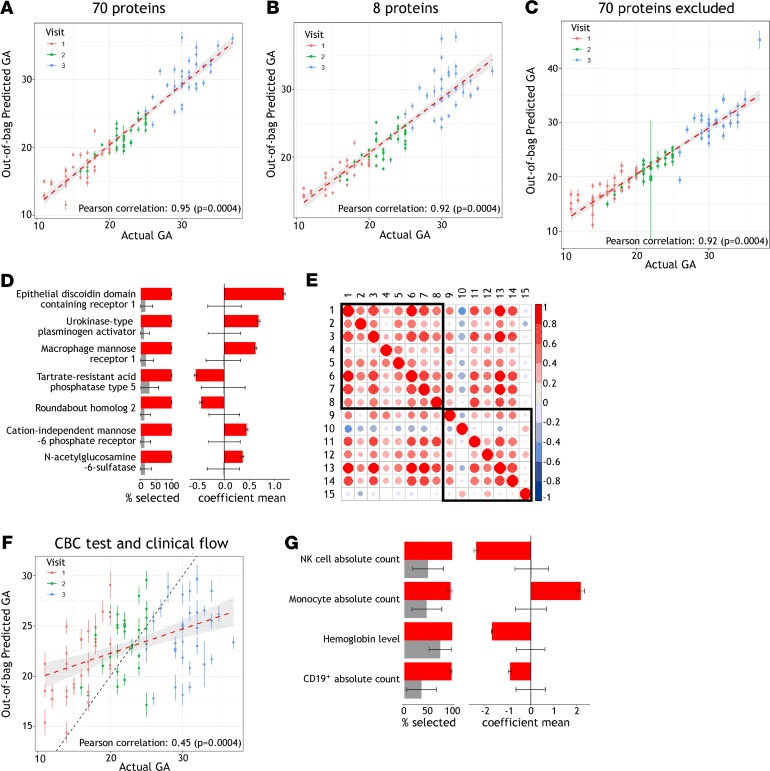
Figure 3. Serum proteins and cell populations predictive of GA.
(A and B) EN models generated from both a set of 70 proteins, and a subset of 8 proteins previously identified to predict GA, demonstrated significant correlation between observed and predicted GA when using serum proteomic data from 3 time points during pregnancy for the 33 women in our study. Predicted GA is shown with mean and standard deviation of 100 model iterations randomly sampling training and test sets, with red dashed line indicating a linear regression with 95% confidence interval. (C) Excluding these 70 proteins and selecting from the remaining 1124 serum proteins measured, EN models still showed significant correlation between observed and predicted GA. (D) For the 7 proteins newly identified to predict GA, frequency of selection and average weights differed significantly from null models and are plotted in red showing mean and standard deviation from the 100 model iterations randomly sampling training and test sets. In gray are shown null models with mean and standard deviation from random permutation of sample labels 25 times and 100 random samplings of the training and test sets. (E) The relationship between new and previously identified proteins predicting GA was assessed by pairwise Pearson’s correlations of protein relative intensities during gestation in our data set. Eight proteins previously identified to predict GA by Aghaeepour et al., and the 7 proteins identified in this study, are numbered 1–8 and 9–15, respectively, and defined as follows: 1, chorionic somatomammotropin hormone; 2, prolactin; 3, macrophage colony-stimulating factor 1 receptor; 4, polymeric immunoglobulin receptor; 5, proto-oncogene tyrosine-protein kinase receptor Ret; 6, glypican-3; 7, granulins; 8, α-fetoprotein; 9, macrophage mannose receptor 1; 10, tartrate-resistant acid phosphatase type 5; 11, N-acetylglucosamine-6-sulfatase; 12, cation-independent mannose-6-phosphate receptor; 13, epithelial discoidin domain-containing receptor 1; 14, urokinase-type plasminogen activator; and 15, roundabout homolog 2. (F) Standard clinical phenotyping measurements from CBC tests and clinical flow cytometry can also be used to predict GA. For the 33 subjects sampled at 3 time points during pregnancy, 19 routinely acquired clinical parameters were used to generate EN models for which predicted and observed GA correlated significantly, with black dashed line marking equal GA. (G) Frequency of selection and average weights are shown for the significant model features, plotted as in D.