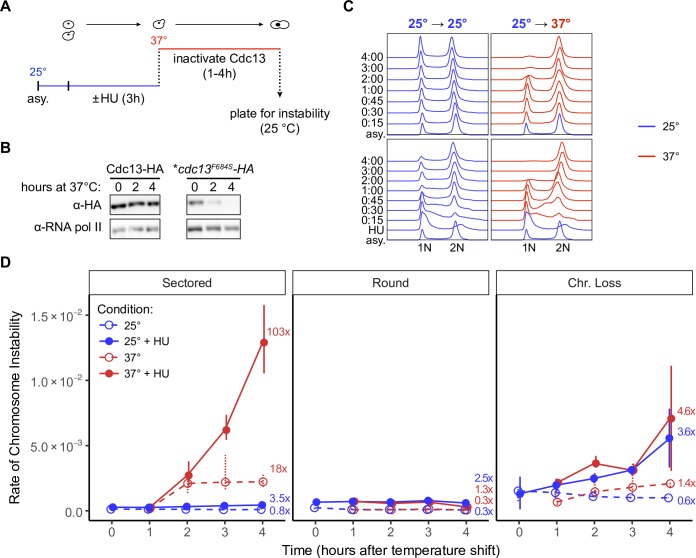
Fig 3. Unstable chromosomes in cdc13F684S form in a single cell cycle.
(A) The experimental protocol for generating unstable chromosomes in a cell cycle. Asynchronous cells were either shifted directly to 37°C or exposed to HU (0.2 M) for 3h at 25°C before the HU was washed out and the cells were shifted to 37°C for 1–4 hours, then plated for instability at 25°C. (B) Exponentially growing Cdc13-HA or cdc13F684S-HA cells were shifted from 25°C to 37°C for 2 or 4h. Whole cell extracts were prepared and analyzed by western blot with an anti-HA antibody. Anti-RNA pol II was used for a loading control. *cdc13F684S-HA also contained the cdc15-2 mutation, further explained in Fig 4. (C) FACS analysis of DNA content. Cells were grown in liquid YPD and prepared as in 2A. After HU arrest 70% of cells were in G1/S phase. The time increments refer to time after HU wash and shift to either 37°C or 25°C. (D) The frequency of sectored, round, and chr. loss colonies in cdc13F684S from 0 to 4 hours after the temperature shift. Blue: incubated at 25°C; red: incubated at 37°C. Fold change and significance are calculated from the 0 hour frequency. Data shown are the medians and IQR of 4 independent experiments.