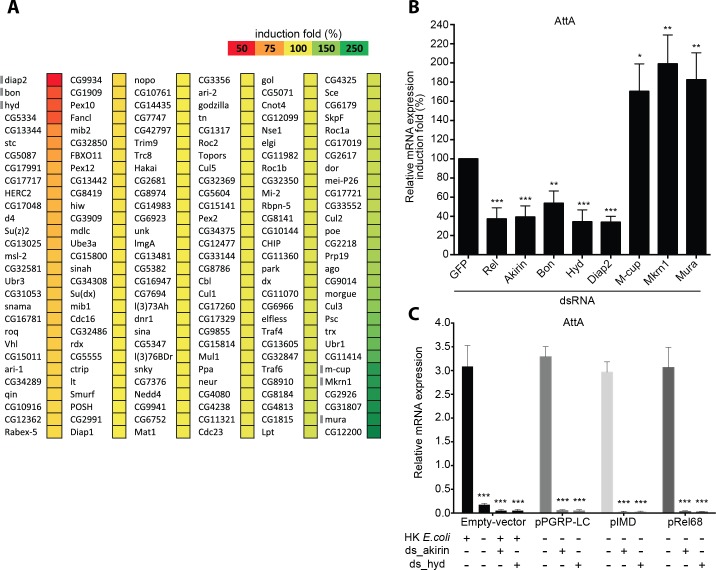
Fig 1. E3-ubiquitin ligases screen identified in vitro Hyd as involved in IMD pathway.
(A) Induction of the IMD pathway measured by luciferase activity. 174 Drosophila E3 ubiquitin ligases were knocked down in S2 cells harboring the Attacin-A–luciferase reporter gene. Cells were transfected with individual dsRNAs targeting each E3 ligase before the IMD pathway was induced by stimulating the cells with heat-killed E. coli (HKE). Luciferase activity is expressed as an induction percentage compared to control cells treated with dsRNA targeting GFP. Three independent experiments were performed. Genes indicated by a gray bar were analyzed in Fig 1B. (B) Quantitative RT-PCR of Attacin-A mRNA from HKE-stimulated S2 cells transfected with dsRNA targeting GFP (negative control), Relish or Akirin (positive controls), and a subset of E3 ubiquitin ligases. After the ratio of stimulated over unstimulated values for each condition was determined, statistical significance was calculated by comparing genes knockdown to GFP dsRNA control. (C) Genetic epistasis experiment to place Hyd within the IMD pathway in S2 cells. The IMD pathway was induced by HKE stimulation or by transfecting the cells with Pgrp-LC, Imd-V5, or Rel-HA expressing plasmids. Cells were also transfected with dsRNA targeting Akirin or Hyd. Statistical significance was established by comparing values from the different conditions with cells treated with empty vector alone. Data are represented as mean ± standard deviation of three independent experiments (B-C). *P-value < 0.05; **P-value < 0.01; ***P-value < 0.001.