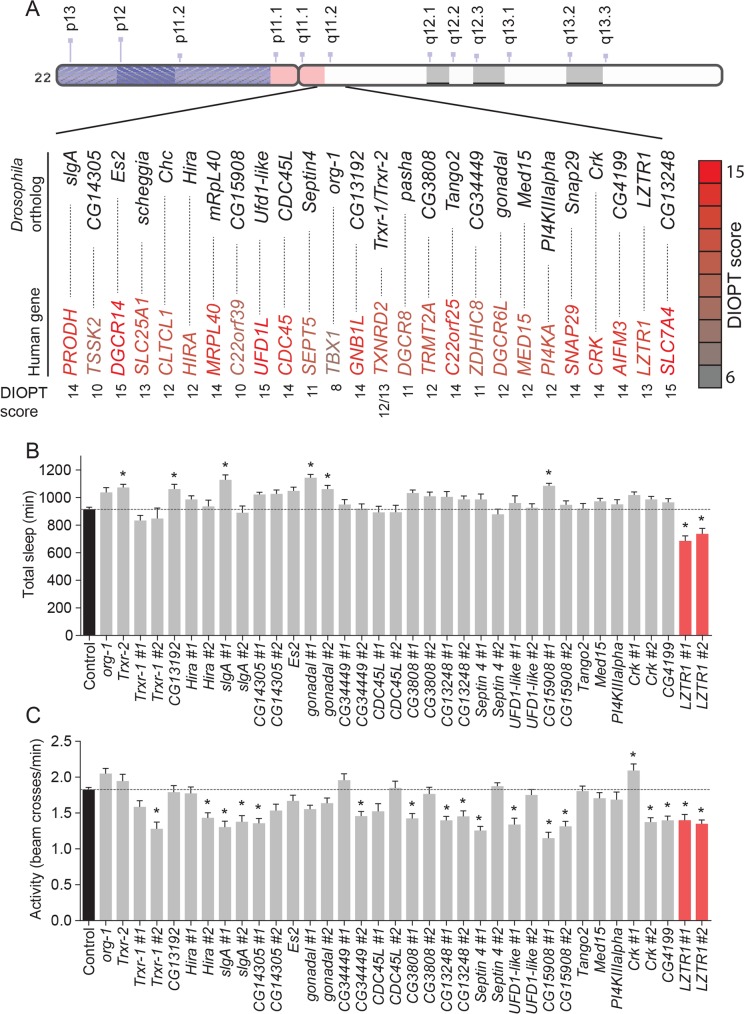
Fig 1. Screening Drosophila orthologs of human genes within the 22q11.2 deletion identifies LZTR1 as a modulator of sleep and activity in Drosophila.
(A) Schematic diagram of the human 22q11.2 deletion. For the 43 genes spanned by the 22q11.2 deletion, 26 orthologs were identified in Drosophila melanogaster with a DIOPT conservation score ≥ 6. (B, C) Total sleep (B) and activity (C) in adult males with pan-neuronal RNAi-mediated knockdown of each of these Drosophila orthologs. The elav-GAL4 (elav>) pan-neuronal driver line was crossed to UAS-RNAi transgenes targeting individual gene orthologs, with n = 16 flies for each genotype. For controls, elav> was crossed to w1118, the genetic background for the RNAi lines, with n = 140 flies. When possible, two independent UAS-RNAi transgenes were used against each gene. Total sleep (minutes) of 3-to-7-day-old males was recorded over a 24-hour period. Transformant ID numbers for RNAi lines are listed in S1 Table. Genes for which knockdown caused lethality or a non-inflating-wing phenotype were not analyzed. Average activity was measured as the number of beam crosses per minute. Knockdown of LZTR1 in the nervous system reduces both total sleep and locomotor activity. Graphs represent means with SEM of data pooled from one to five independent experiments. Significance was determined using Kruskal-Wallis test with Dunn's post-hoc testing (* p<0.05).