Figure 5. hRpn2-bound hRpn13 binds to an extended conformational of K48-diubiquitin.
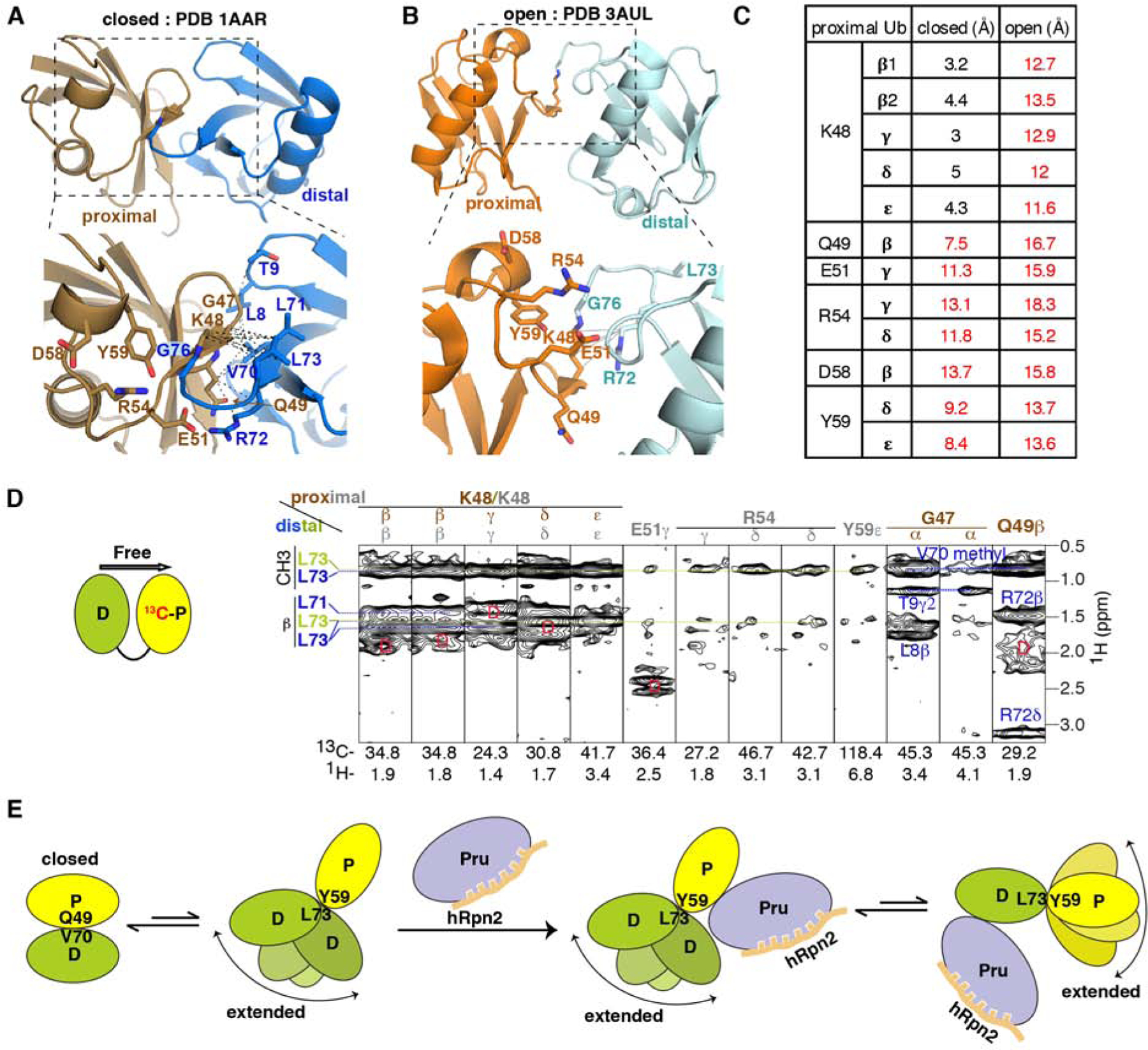
(A and B) Ribbon diagram of free K48-diubiquitin structures solved by x-ray crystallography with an expansion (dashed rectangles) in the lower panels. Two conformational states have been reported and named ‘closed’ (PDB 1AAR, A, proximal in brown, distal in blue) or ‘open’ (PDB 3AUL, B, proximal in orange and distal in cyan). Sidechains of interacting ubiquitin residues are displayed (bottom panels), with nitrogen and oxygen in blue and red, respectively. Black dashed lines represent inter-ubiquitin NOE interactions detected in the experiment of (D), involving proximal ubiquitin G47-Q49 and distal ubiquitin L8-T9 or V70-L73 (A). Gray solid lines represent interactions close enough for expected NOE interactions between proximal ubiquitin K48 and distal ubiquitin R72 (B); these were not detected in our 13C-half-filtered NOESY experiment.
(C) Measured distances between proximal ubiquitin residues and distal ubiquitin L73 for the ‘closed’ (Figure 5A) and ‘open’ (Figure 5B) crystal structures of free K48-diubiquitin. Distances too large for NOE interactions are indicated in red.
(D) Selected regions from a 13C-half-filtered NOESY experiment (100 ms mixing time, right panel) acquired on 0.3 mM K48-diubiquitin with 13C-labeled proximal (left panel, yellow) and unlabeled distal (left panel, green) ubiquitin at 25°C and pH 6.5 highlighting NOE interactions between the two ubiquitin moieties. NOE interactions expected from the ‘closed’ conformation are labeled brown for proximal ubiquitin and blue for distal ubiquitin whereas those consistent with the structure used to bind hRpn13 (Figures 1C–1F) are labeled gray for proximal ubiquitin and green for distal ubiquitin. Breakthrough diagonal signals are indicated with a red ‘D’.
(E) Cartoon depicting the dynamics of free K48-diubiquitin and following interaction with hRpn2-bound hRpn13. When free, K48-diubiquitin exchanges between the ‘closed’ conformation resolved by x-ray crystallography (A) and an ensemble of extended conformations, as detected in this study. The ‘closed’ conformation is dominant for free K48-diubiquitin. The extended conformation is selected by hRpn2-bound hRpn13, which binds either ubiquitin with preference for the proximal moiety. See also Figure S4.
