Figure 6. Comparison with previous structures of hRpn13 complexed with ubiquitin.
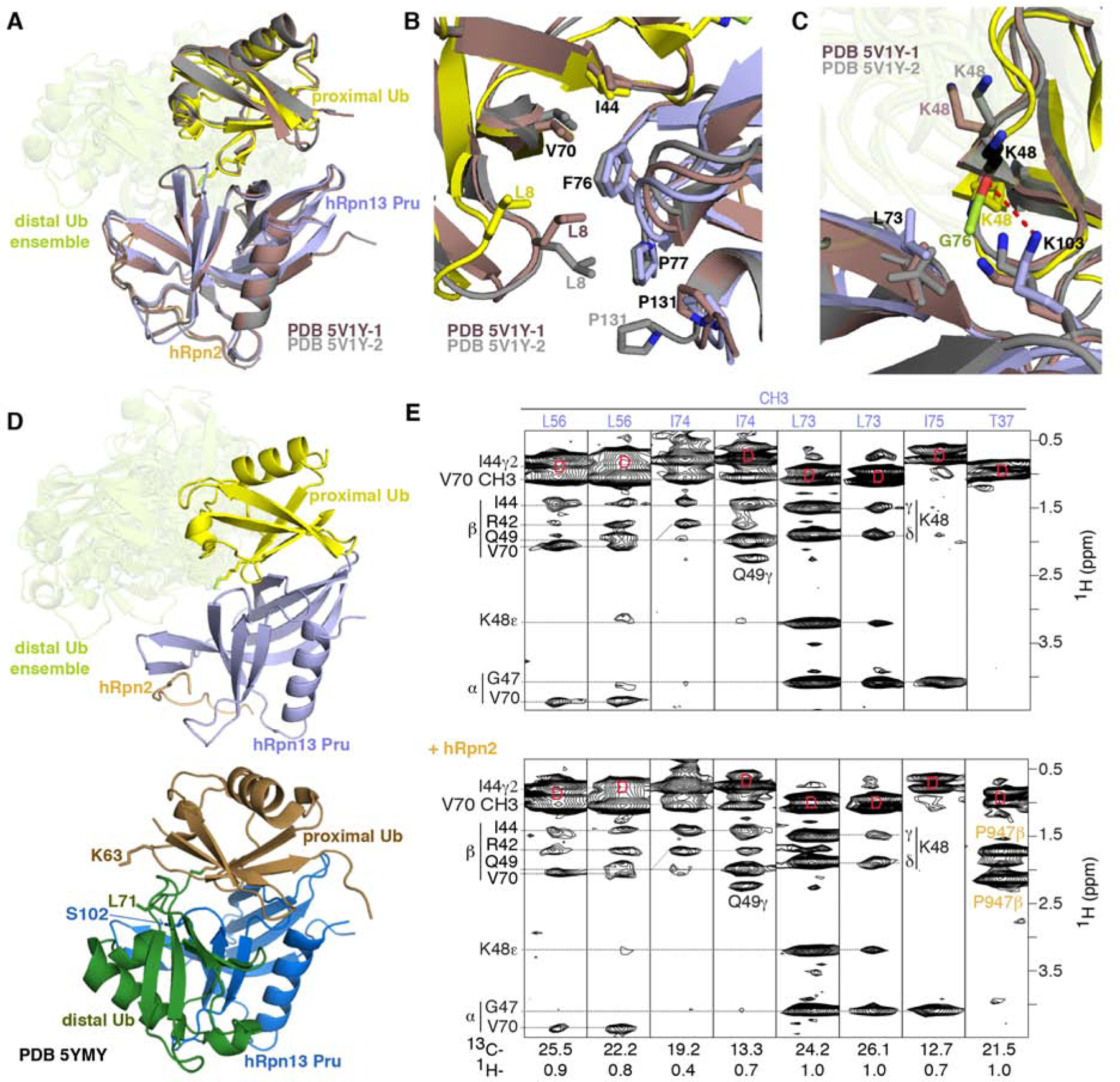
(A, B and C) Superposition of hRpn13 Pru:hRpn2 (940–953):ubiquitin for the two available crystal forms (PDB 5V1Y-1 in dark red and PDB 5V1Y-2 in gray) and our Ternary-P structure presented as in Figure 1E. Expanded regions centered on ubiquitin L8-I44-V70 (B) and ubiquitin K48 (C) are included. Interactions to hRpn13 F76, P77, and P131 are highlighted in (B) demonstrating similarity among the three structures for hRpn13 F76 but divergence for ubiquitin L8 and hRpn13 P131. In the crystal structures with monoubiquitin, K48 adopts two configurations (displayed in grey and black) for PDB 5V1Y-2 and one conformation for PDB 5V1Y-1 (dark red). The isopeptide bonded ubiquitin K48 is directed closer to L73 compared to in the structures with monoubiquitin (C). From this location, a hydrogen bond (red dashed line) forms between the hRpn13 K103 ε-ammonium group and the carbonyl of isopeptide bonded ubiquitin G76. Nitrogen and oxygen atoms are displayed in blue and red, respectively.
(D) Structural comparison for Ternary-P (top panel, displayed as in A) and structure PDB-5YMY of hRpn13 Pru (blue):K48-diubiquitin (bottom panel, proximal or distal ubiquitin in brown or dark green, respectively) with identical orientation for hRpn13 Pru. In PDB-5YMY, hRpn13 S102, proximal ubiquitin K63 and distal ubiquitin L71 are displayed and labeled.
(E) Selected regions of a 13C-half-filtered NOESY experiment (mixing time 100 ms) acquired on a mixture of 0.25 mM 13C-labeled hRpn13 Pru with 1.2-fold molar excess unlabeled monoubiquitin without (top panel) or with 1.2-fold molar excess hRpn2 (940–953) (bottom panel) at 25°C and pH 6.5. Diagonal br eakthrough signals are labeled by ‘D’ (red) and assignments for hRpn13 Pru (purple), ubiquitin (black) and hRpn2 (orange) included. See also Figure S3.
