Figure 7. . Model illustrating how dynamic interactions between ubiquitin chains and proteasome receptors may stochastically drive substrate proteolysis.
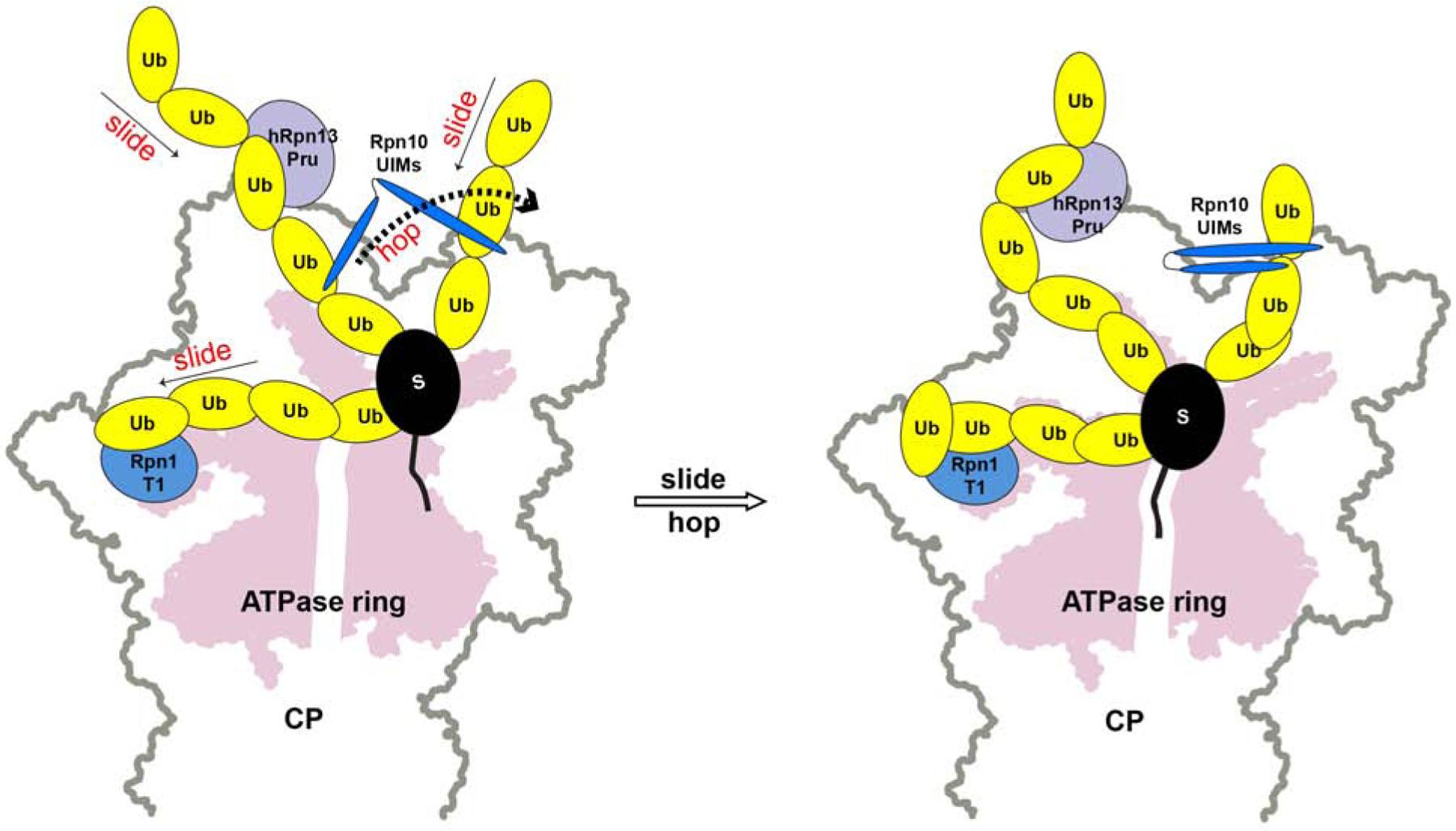
Based on our previous and current NMR data, we propose that the three substrate receptors hRpn1 (blue), hRpn10 (dark blue) and hRpn13 (purple) bind dynamically to ubiquitin chains (yellow) attached to substrate (black), sliding within a chain and hopping between chains, causing motion for the substrate (left panel) until a productive orientation is formed whereby the substrate engages the ATPase ring (pink, right panel).
