Extended Data Fig. 9. Transcription factors that preferentially bind at ssDNA-containing enhancers in HEK293T cells.
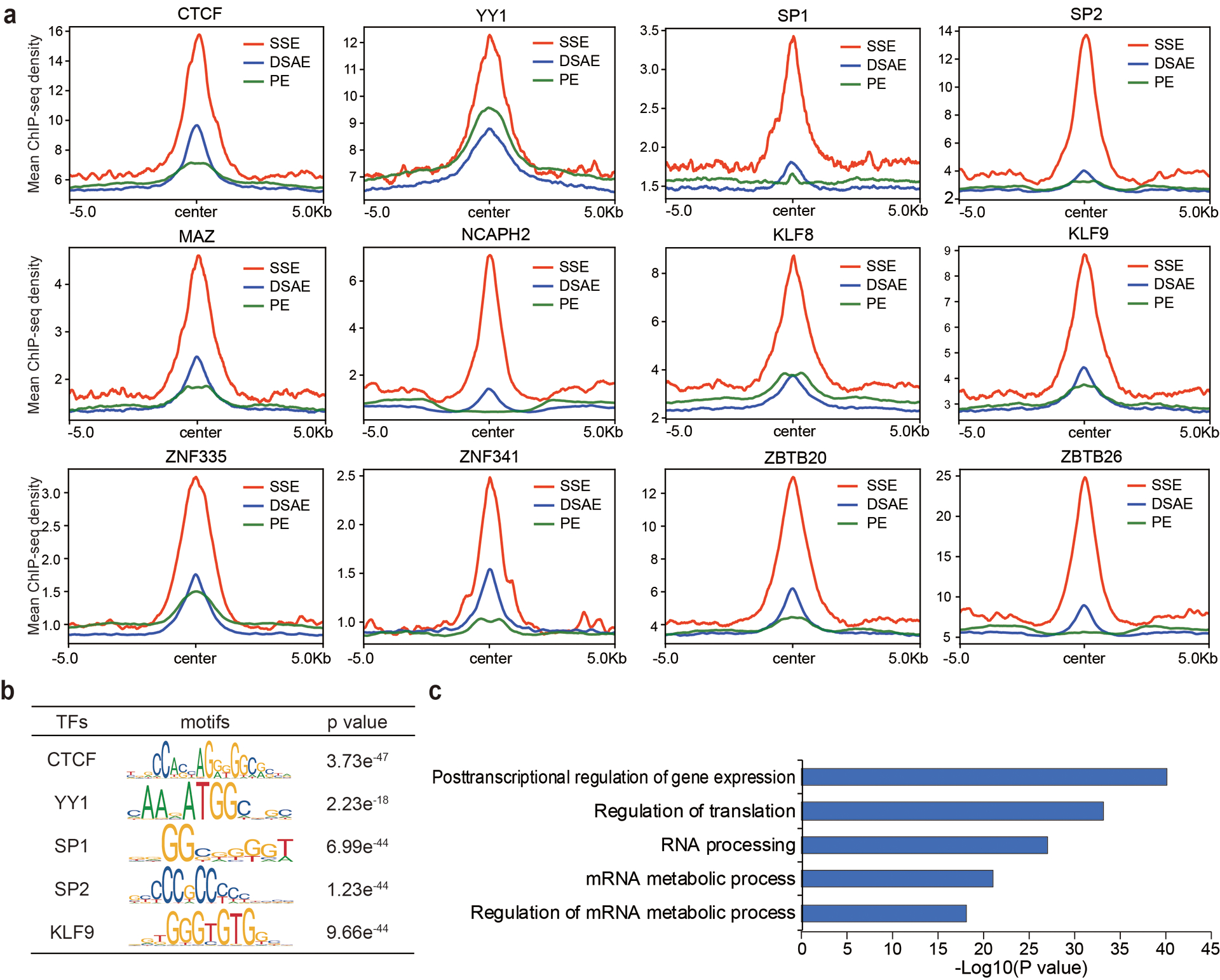
a, Metagene profiles of CTCF, YY1, SP1, SP2, MAZ, NCAPH2, KLF8, KLF9, ZNF335, ZNF341, ZBTB20, and ZBTB26 ChIP-seq reads densities at denoted enhancers in HEK293T cells. Regions within 10 kb around the enhancer centers are shown. b, Transcription factor binding motifs enriched at ssDNA-containing enhancers (n = 1,969 enhancers) in HEK293T cells with corresponding p values by using the genome as background. Only TFs with motif information in the TRANSFAC vertebrates library were analyzed. P values were calculated by two-sided binomial test. c, GREAT analysis of genes regulated by ssDNA-containing enhancers (n = 1,969 enhancers) in HEK293T cells. P values were calculated by two-sided binomial test. SSE: ssDNA-containing enhancers; DSAE: double-stranded active enhancers; PE: poised enhancers.
