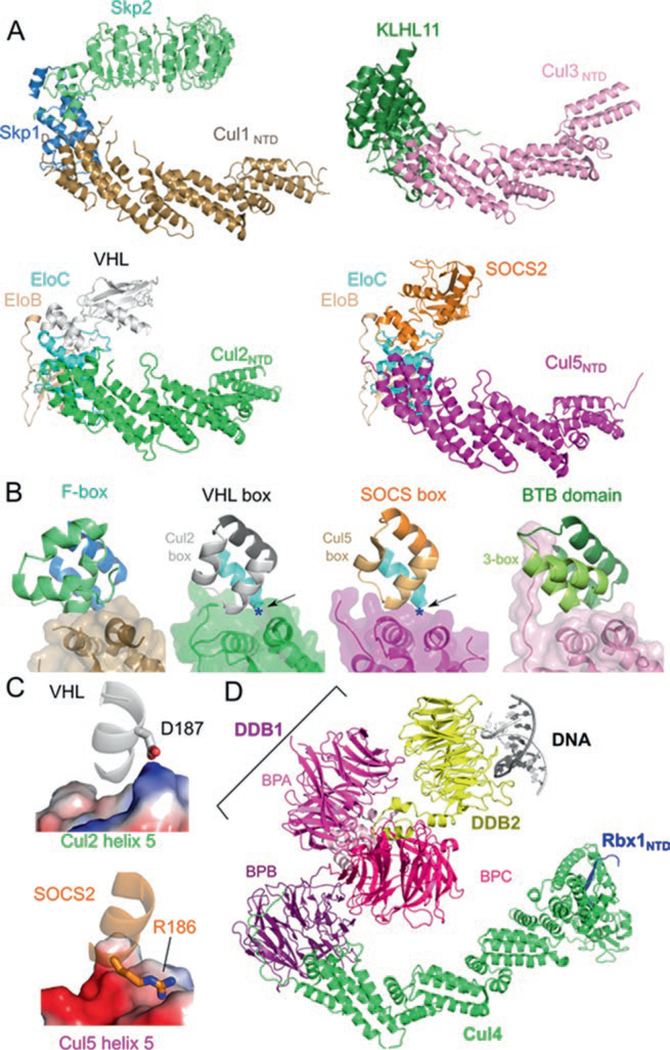
Fig. 12.2.
Similar architecture of cullin-RING ligases. (a) Structures of VHL–EloBC–Cul2NTD (PDB ID: 4WQO), SOCS2–EloBC–Cul5NTD (PDB ID: 4JGH), Skp2–Skp1–Cul1NTD (PDB ID: 1LDK), and KLHL11-Cul3NTD (PDB ID: 4AP2) show a common spatial organization of components. Cullin repeats 2 and 3 of Cul2 were modeled from Cul1. (b) The cullin–adaptor–substrate receptor binding regions for each CRL shown in (a) are in ribbon representations with semitrans-parent surfaces shown for the cullins. The Cul2 box, the Cul5 box, and the 3-box are marked. (c) Charge complementary at the binary cullin–substrate receptor interface between VHL and Cul2 helix α5 (top) or between SOCS2 and Cul5 helix α5 (bottom). Cullins are shown in electrostatic surface representation (Blue: positive; Red: negative). Each region is marked in (b) with a blue asterisk and with the point of view depicted by the arrow. (d) Structure of the CRL4DDB2–UV-damaged DNA complex (PDB ID: 4A0K) in ribbon representation. The domains of DDB1 are marked