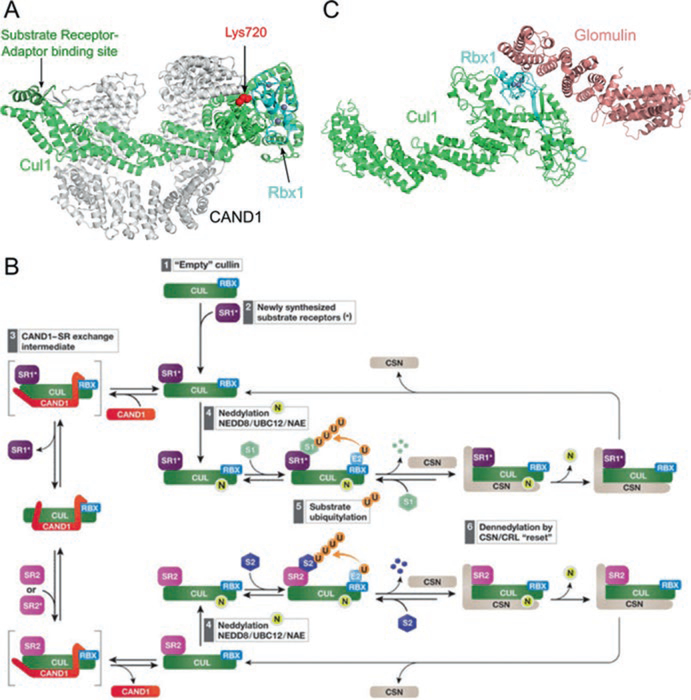
Fig. 12.4.
Regulation of cullin-RING ligases by CAND1 and Glomulin. (a) Structure of Cul1–Rbx1 inhibited by CAND1 (PDB ID: 1U6G) in ribbon representation. CAND1 sterically blocks the neddylation site on the cullin (shown in red spheres) and the substrate receptor binding site at the cullin NTD (highlighted in dark green). (b) Model of remodeling of CRL composition by CAND1 and CSN (Reprinted from Lydeard et al. 2013. See main text for description). (c) Structural inhibition of Rbx1 by glomulin (PDB ID: 4F52) show in ribbon representation. Glomulin inhibits CRL activity by binding to Rbx1 and blocking the E2 interacting site. The Zn2+ ions in (a) and (c) are shown as gray spheres