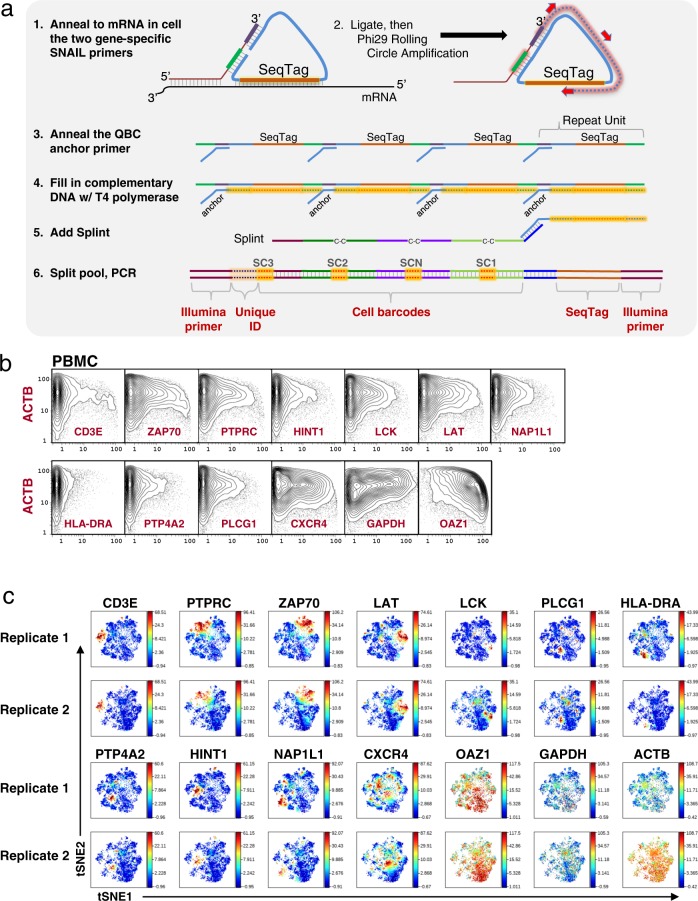
Fig. 10. In situ amplification.
a Schematic for target specific in situ amplification by SNAIL padlock amplification. (1) Sets of primer pairs are added to fixed cells and allowed to anneal to target mRNA. Padlock mediated ligation is performed. (2) Rolling circle amplification of the circle amplifies the sequence tag and a region complementary to the QBC anchor primer. (3) The anchor primer is added and (4) T4 polymerase extends the primer. (5) Splint is added and (6) Split-pool as per prior versions of the approach for protein and mRNA, PCR, sequencing, and data analysis. b Single-cell expression analysis of 14 mRNA transcripts in primary PBMC cells. Human PBMCs were prepared for SNAIL amplification against 14 mRNA transcripts. Plots represent 21,143 cells. Bi-axial plots against ACTB for all transcripts are shown. c Expression of T cell, B cell, and housekeeping RNA transcripts demonstrated via viSNE plots. Data from two SNAIL biological replicates were rendered via viSNE using HLA-DRA, CXCR4, HINT1, CD3E, ZAP70, LCK, LAT, PTP4A2, PTPRC, PLCG1, and NAP1L1 as selected channels to organize the viSNE plots. The housekeeping markers GAPDH, ACTB, and OAZ1 were deliberately excluded from the viSNE run to view how the expression of the other markers correlated with each other. Expression of the RNA targets varied across the cells. CD3E and HLA-DRA are known to be anti-expressed and their observed expression patterns are in non-overlapping regions of the viSNE plots.