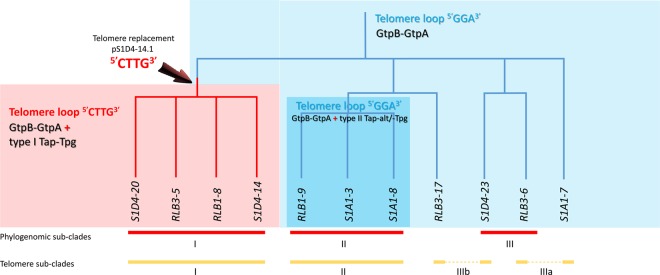
Figure 1.
Telomere and terminal protein distribution among the Streptomyces population. A maximum likelihood phylogenomic tree of the 11 strains was built based on 5,149,602 nucleotide positions. Heavy lines indicate branches supported by bootstrap values >80% (100 replicates). According to the tree topology and bootstrap values, three phylogenomic sub-clades (I, II and III) were defined. The strains belonging to a same phylogenomic sub-clade are connected with a red bar. The yellow bars connect stains harbouring identical or highly closely related telomere sequences (more than 92% sequence identity). The dashed yellow lines indicate incongruences between the phylogenomic tree and the distribution of similar telomeres. The coloured backgrounds represent the distribution of the chromosomal telomere loops and of the different tap-tpg/gtpB-gtpA systems. The light blue background encompasses strains for which only the gtpB-gtpA system was identified. Their chromosomal telomeres are typified with a 5′GGA3′ loop. A parsimonious evolutionary scenario suggest that the ancestor of the population shared the same combination. The turquoise background encompasses strains having also telomeres with a 5′GGA3′ loop and a gtpB-gtpA operon, but with an additional type II tap-tpg operon. The light red background represents strains having the ancestral gtpB-gtpA operon, but also an additional type I tap-tpg operon and telomeres typified by a 5′CTTG3′ loop. The red arrow indicates a potential telomere replacement with a plasmid at the root of this sub-cluster.