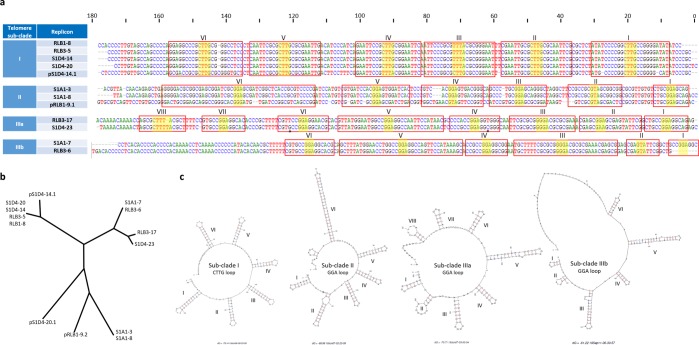
Figure 5.
Comparison of the different chromosomal and plasmidic telomeres. (a) The terminal 180 nucleotide sequences from the 10 chromosomes and 2 linear plasmids were aligned. Four different groups designed in the table as telomere sub-clades were defined according to their sequence identities. Palindromic sequences are boxed and numbered in Arabic numerals above the sequences and the telomere loops are highlighted in yellow. Compensatory nucleotide changes within palindromes IV and VI of RLB3-7 telomere are highlighted in blue. The telomere sequence of plasmid pS1D4-20.1 could not be aligned with the others and is not presented. (b) Unrooted NJ phylogenetic tree built with the different telomere sequences. Positions with <80% site coverage in the alignment were eliminated enabling to have a total of 164 nucleotide positions in the final dataset. Bootstrap percentages are indicated on the branches. (c) The predicted secondary structures for a representative sequence of each telomere sub-cluster are represented. Two different loops (CTTG and GGA) can be observed at the top of the hairpin structures.