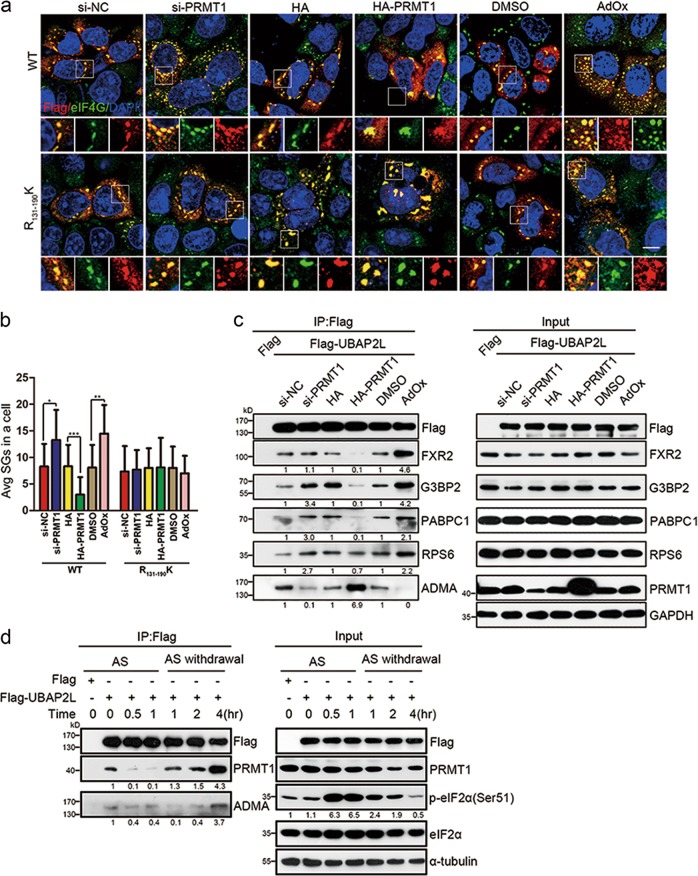
Fig. 6.
PRMT1 governs arginine methylation of UBAP2L to monitor SG dynamics. a SG formation in UBAP2L-KD HeLa cells expressing the Flag-tagged UBAP2L-WT or UBAP2L-R131–190K mutants with the indicated treatments. UBAP2L-KD HeLa cells were transfected with PRMT1-siRNA or HA-PRMT1 plasmid, or treated with Adox, adenosine dialdehyde, a pan methyltransferase inhibitor (20 μM, 24 h), and then stressed in AS (500 μM, 1 h), using Flag (red)/eIF4G (green) as SG markers. Scale bars = 10 μm. b Quantification of SGs in (a). Bars indicate the average number of SGs in Flag-positive cells, and differences between groups were analyzed using the t-test. **P < 0.01, n = 3. c Co-IP/WB analysis of the associations of UBAP2L with SG proteins in HEK293 cells transfected with PRMT1-siRNA or HA-PRMT1 plasmid, or treated with Adox. d UBAP2L association with PRMT1 and its ADMA level through stress and recovery periods. HEK293 cells expressing Flag or Flag-UBAP2L-WT were stressed with 500 μM AS for 1 h, and then recovered for indicated periods. PRMT1 and ADMA levels were determined by immunoblotting analysis of Flag precipitates. p-eIF2α (S41) in the lysates was shown as a proxy for translation inhibition during the stress and recovery periods