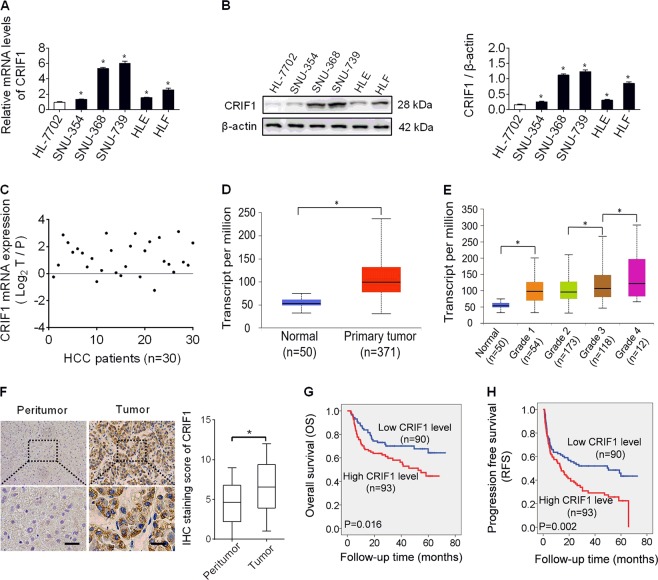
Fig. 1. CRIF1 was frequently overexpressed in HCC and contributes to worse prognosis.
a Quantitative real-time PCR (qRT-PCR) analysis for mRNA expression levels of CRIF1 in five human HCC cell lines (SNU-354, SNU-368, SNU-739, HLE, and HLF) and one normal human hepatic cell line HL-7702. b Western blot analysis for protein expression levels of CRIF1 in five human HCC cell lines (SNU-354, SNU-368, SNU-739, HLE, and HLF) and one normal human hepatic cell line HL-7702. c Quantitative RT-PCR analysis for mRNA expression levels of CRIF1 in paired tumor and peritumor tissues from 30 HCC patients. (T tumor, P peritumor). d, e UALCAN (a portal for facilitating tumor subgroup gene expression and survival analyses) websites-based bioinformatic analysis of CRIF1 expression in HCC. f Immunohistochemical staining for expression levels of CRIF1 in paired tumor and peritumor tissues from 183 HCC patients. Left panel: representative immunohistochemical (IHC) staining images of CRIF1 in paired tumor and peritumor tissues of HCC. Scale bar, 50 μm. Right panel: IHC staining scores of CRIF1 in paired tumor and peritumor tissues of HCC. g, h Overall survival (OS) and recurrence-free survival (RFS) were evaluated by the Kaplan–Meier survival curves and the log-rank test in tumor tissues from 183 HCC patients according to CRIF1 expression levels. Data are expressed as mean ± SEM from three independent experiments. *p < 0.05.