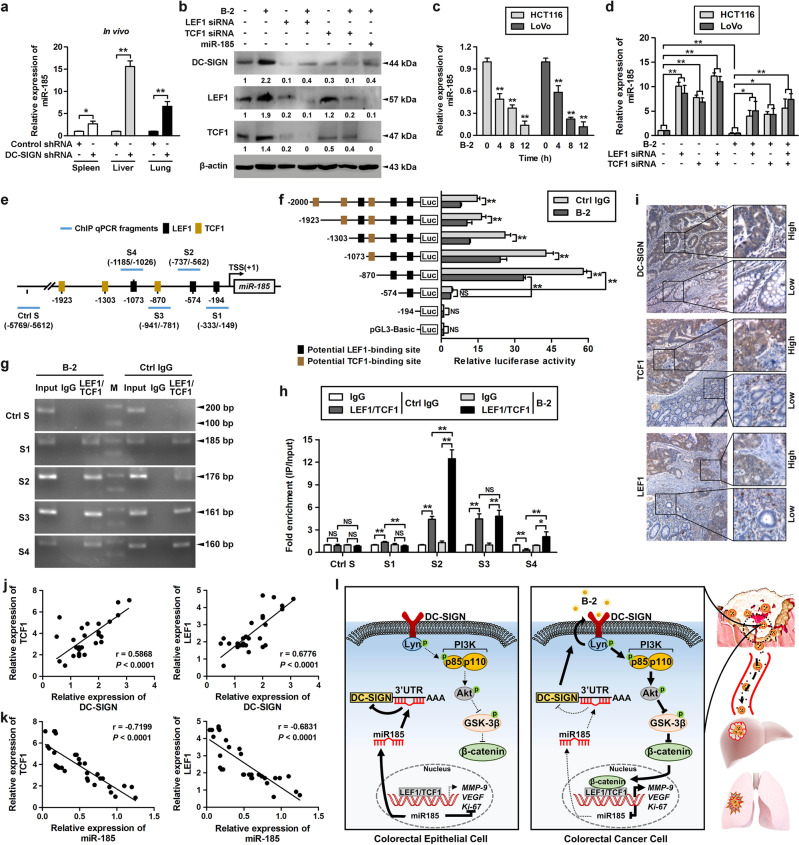
Fig. 6.
β-catenin/TCF1/LEF1 directly suppresses miR-185 expression in CRC cells. a Stable LoVo cells expressing control shRNA or DC-SIGN shRNA were injected into the spleen. Then total RNA of spleen, lung, and liver tissues was extracted and applied to qPCR. b Cells were transfected as indicated and treated with DC-SIGN mAb (B-2, 10 μg/ml), followed by western blot. c Cells were treated with DC-SIGN mAb, and expression of miR-185 was examined using qPCR. d Cells infected with LEF1 and/or TCF1 siRNA were treated with DC-SIGN mAb, and total RNA was extracted and applied to qPCR. e Schematic presentation of the genomic localization of the human miR-185 gene and its promoter region. f Serially truncated miR-185 promoter constructs were cloned to pGL3-luciferase reporter plasmids and transfected into LoVo cells. Cells were treated with DC-SIGN mAb, and applied in luciferase activities analysis. g Chromatin of LoVo cells cultured in the absence and presence of DC-SIGN mAb was subjected to chromatin immunoprecipitation with antibodies against LEF1 or TCF1 followed by PCR analysis using primers amplifying different regions of the miR-185 promoter indicated in d. M, DL1000 Marker. h qPCR of the ChIP products validated the binding capacity of LEF1 and TCF1 to the miR-185 promoter. i Representative images of IHC staining for DC-SIGN, LEF1, and TCF1 in CRC tissue. j, k Correlation between LEF1 or TCF1 expression and DC-SIGN (j) or miR-185 (k) levels in CRC samples. Spearman correlation coefficient with the respective significance is indicated. l Schematic representation of the proposed roles of DC-SIGN–LEF1/TCF1–miR-185 feedback loop in the of CRC progression. Data, mean ± SD. *P < 0.05, **P < 0.01. N.S., nonsignificant