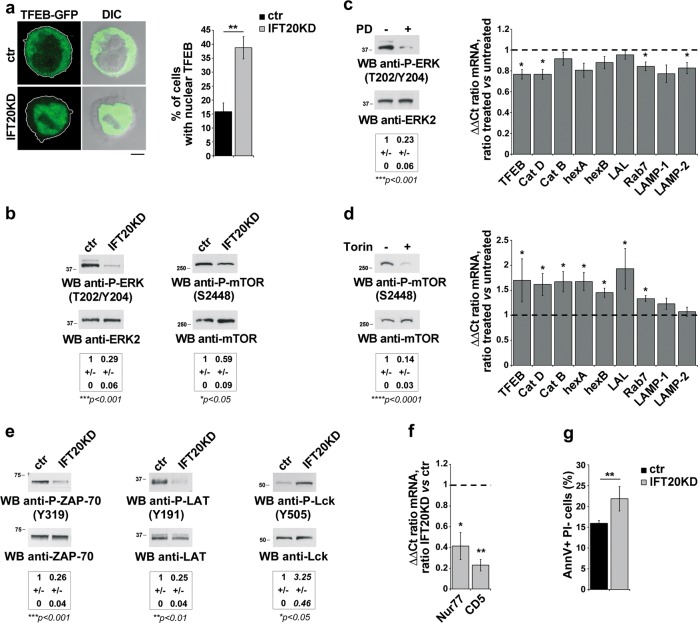
Fig. 4.
IFT20 couples tonic TCR signaling to TFEB activation. a Quantification of nuclear GFP-tagged TFEB in ctr and IFT20KD Jurkat cells (mean ± SD; n = 3; Student’s t test). Representative images (medial optical sections) are shown. b Immunoblot analysis with anti-P-ERK1/2 or P-mTOR antibody of lysates from ctr and IFT20KD Jurkat cells. ERK2 or mTOR was used as loading control. The quantification of the relative P-protein levels, normalized to the respective loading controls, is reported below (mean fold ± SD; ctr value = 1; n ≥ 3; one sample t test). c Left, Immunoblot analysis with anti-P-ERK1/2 antibody of lysates from Jurkat cells, treated for 16 h with 20 μM PD098059. ERK2 was used as loading control. Right, Quantitative RT-PCR analysis of TFEB-regulated genes and TFEB in PD098059-treated ctr Jurkat T cells. The relative abundance of gene transcripts was determined on samples using the ddCt method and was normalized to HPRT1. The data (mean ± SD; n ≥ 3; one sample t test) are expressed as normalized fold expression in treated vs untreated cells (expression in untreated cells set for each gene as 1, dashed line). d Left, Immunoblot analysis with anti-P-mTOR antibody of lysates from Jurkat cells, treated for 16 h with 250 nM Torin1. mTOR was used as loading control. Right, Quantitative RT-PCR analysis of TFEB-regulated genes and TFEB in Torin1-treated ctr Jurkat T cells. The relative abundance of gene transcripts was determined on samples using the ddCt method and was normalized to HPRT1. The data (mean ± SD; n ≥ 3; one sample t test) are expressed as normalized fold expression in treated vs untreated cells (expression in untreated cells set for each gene as 1, dashed line). e Immunoblot analysis with anti-P-ZAP-70 (Y319), anti-P-LAT (Y191) or anti-P-Lck (Y505) antibody of lysates from ctr and IFT20KD Jurkat cells. ZAP-70, LAT or Lck was used as loading control. The migration of molecular mass markers is indicated. The quantification of the relative P-protein levels, normalized to the respective loading controls, is reported below (mean fold ± SD; ctr value = 1; n = 3; one sample t test). f Quantitative RT-PCR analysis of Nur77 and CD5 expression in ctr and IFT20KD Jurkat cells. The relative abundance of gene transcripts was determined on triplicate samples using the ddCt method and was normalized to HPRT1. The data (mean ± SD; n = 3) are expressed as normalized fold expression in IFT20KD vs control cells (expression in ctr cells set for each gene as 1, dashed line) (one sample t test). g Flow-cytometric analysis of apoptosis in ctr and IFT20KD Jurkat cells grown for 24 h in the culture medium supplemented with 0.5% BCS and labeled with Annexin V and propidium iodide. The histograms show the percentages of Annexin V-positive propidium iodide-negative cells (mean ± SD; n = 6; Student’s t test). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001