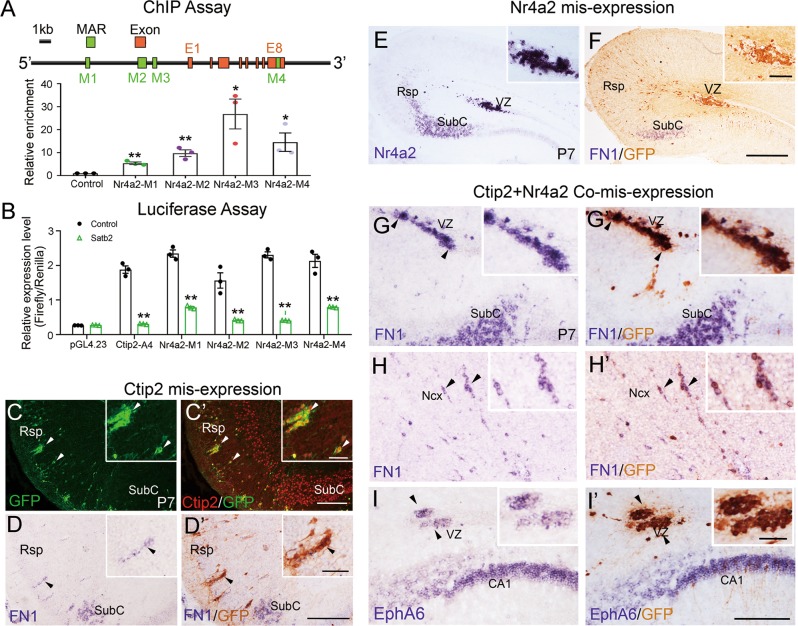
Fig. 6.
Satb2 determines Rsp fate by repressing Ctip2 and Nr4a2. a Diagram of Satb2-binding MARs in Nr4a2 genomic locus. In vivo ChIP assay using embryonic mouse cortex shows a remarkable enrichment of Satb2-binding MARs in Nr4a2 locus. *p < 0.05, **p < 0.01 (n = 3), error bars represent S.E.M. b Luciferase reporter assay shows that the transcriptional activities of Ctip2-A4 (positive control) and four Nr4a2 MARs are significantly reduced in the presence of Satb2. **p < 0.01 (n = 3), error bars represent S.E.M. c–i′ Ctip2- and/or Nr4a2-expression plasmids together with GFP are delivered into Rsp neurons by in utero electroporation in wild-type mice at E14.5 and pups are analyzed at P7. To make signals more clearly to compare, mRNA signals were photographed first, and then immunostaining of GFP was performed and photographed again. c, c′ Ctip2 is misexpressed in wild-type Rsp, and some of them are clumped. Inserts show Ctip2 expression in GFP+ cells (arrowheads). d, d′ A few Ctip2-misexpressed neurons contain FN1 transcripts and they often reside in ventral Rsp (arrowheads). Most of Rsp neurons with misexpression of Nr4a2 are arrested in VZ (e), and none of them contains FN1 transcripts in wild-type Rsp (f). Rsp neurons with co-misexpression of Ctip2 and Nr4a2 initiate FN1 expression in the VZ of Rsp (g, g′) and neocortical region (h, h′). i, i′ EphA6 is also ectopically induced in wild-type Rsp neurons when Ctip2 and Nr4a2 are co-misexpressed. VZ ventricular zone; Ncx neocortex. n = 3 mice for each group. Scale bars = 200 μm in c′, d′, and i′, 50 μm in inserts of c′, d′, and i′, 400 μm in f, and 100 μm in inserts of f