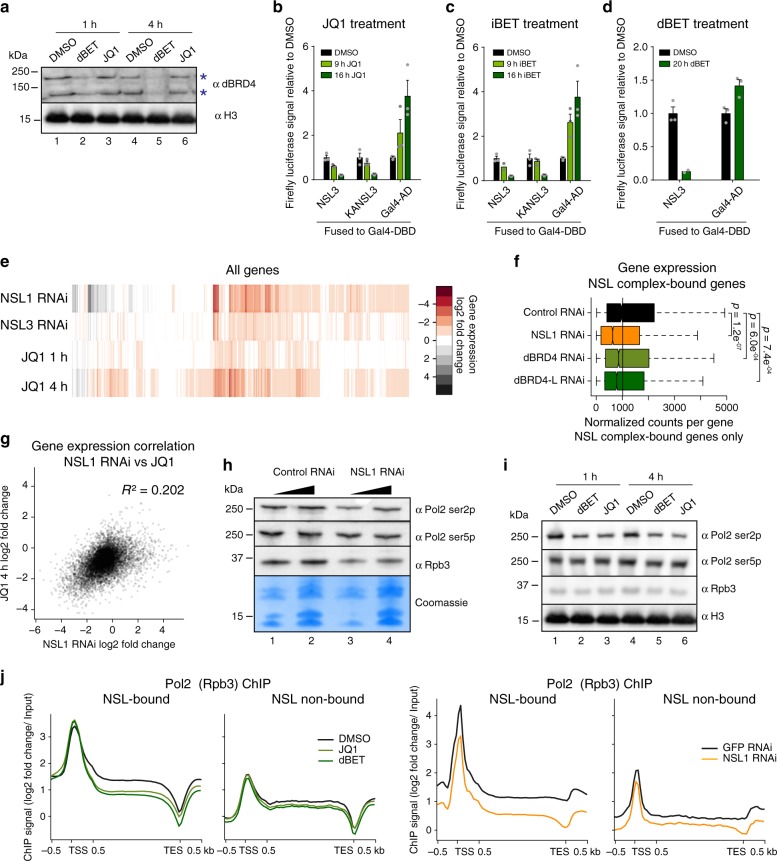
Fig. 3. dBRD4 promotes transcription elongation of NSL target genes.
a Western blot for dBRD4 and H3 after 100 nM dBET6 (lanes 2 and 5) or 5 µM JQ1 (lanes 3 and 6) treatment. Blue asterisks indicate dBRD4-S and dBRD4-L. The experiment was repeated twice showing similar results. b–d Firefly luciferase activity using NSL3 and KANSL3 (human orthologue of NSL3) fused to Gal4 DNA-binding domain or full-length Gal4 to drive expression of the UAS-firefly luciferase reporter upon (b) 5 µM JQ1, (c) 1 µM iBET 762 or (d) 100 nM dBET6 treatment. Bars represent mean values ± SEM (n = 3 technical replicates). e Heatmap of total RNA-seq. Log2 fold changes of gene expression in NSL1 and NSL3 RNAi and JQ1 (5 µM) treatments for 1 h or 4 h versus control RNAi (GST) or DMSO for all expressed genes are plotted. Gene order was generated by unsupervised hierarchical clustering. f Boxplot of normalized RNA-seq counts in NSL1, dBRD4, dBRD4-L and control RNAi (GST) for NSL complex-bound genes (n = 5600). Two-sided Welch two sample t-test was applied. Boxplots show median (centre), interquartile-range (box) and minima/maxima (whiskers). g Scatterplot of gene expression changes after 4 h JQ1 treatment and NSL1 RNAi. Log2 fold changes for all genes are plotted. Linear regression model was applied. h Representative western blot for Pol2 ser2p, Pol2 ser5p and Rpb3 after NSL1 RNAi (lanes 3 and 4), for quantification see Supplementary Fig. 3j. H4 blot shown here is identical to a. i Representative western blot for Pol2 ser2p, Pol2 ser5p, Rpb3 and H3 after 100 nM dBET6 (lanes 2 and 5) or 5 µM JQ1 (lanes 3 and 6) treatment, for quantification see Supplementary Figure 3j. j. Average profiles of Pol2 (Rpb3) ChIP-seq signal for expressed NSL-bound (n = 5600) and expressed non-NSL-bound (n = 1600) genes after 1 h JQ1 (5 µM), 1 h dBET6 (100 nM) or NSL1 RNAi13 compared to controls (DMSO and GFP RNAi). Gene bodies are scaled from 0.5 kb until TESs. Drosophila virilis chromatin was added to experimental Drosophila melanogaster chromatin before Rpb3 ChIPs to control for IP efficiency and library composition effects (see Methods). e–g Expression was normalized using synthetic ERCC spikes (see Methods). n = 3 biological replicates. Source data for b–d are provided as a Source Data file.