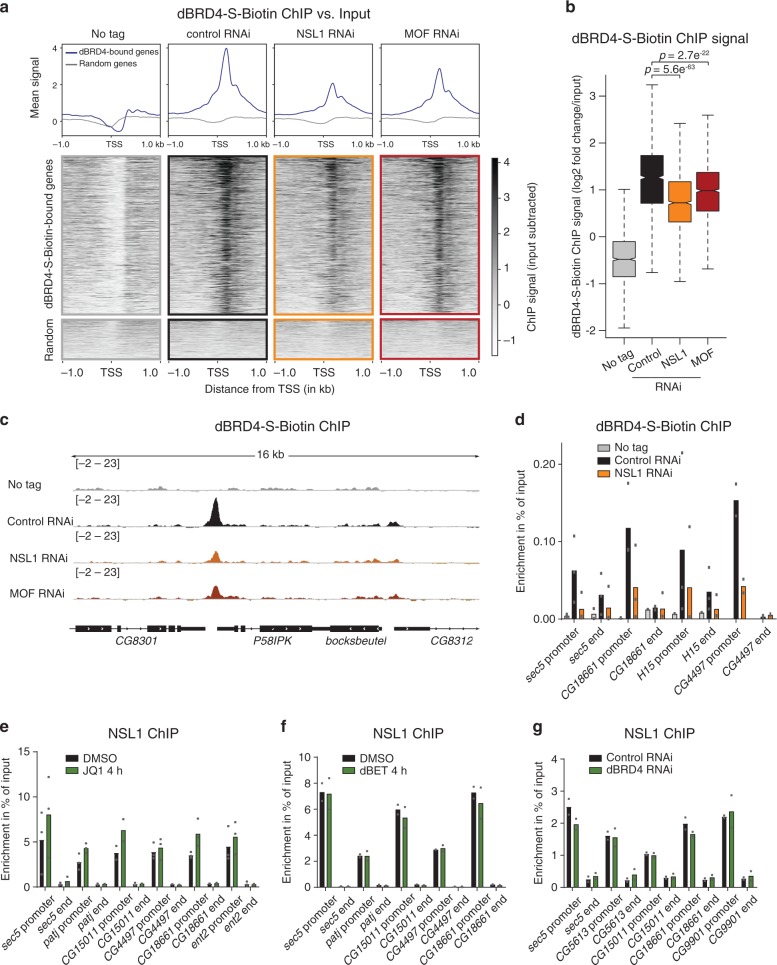
Fig. 4. Efficient recruitment of dBRD4 requires the NSL complex.
Experiments addressing dBRD4 targeting were performed in a stable S2 cell line, where the short isoform of dBRD4 triple tagged with FLAG, His and biotin (dBRD4-S-Biotin) was under the control of a copper-inducible promoter. For all experiments with this stable cell line, expression of dBRD4-S-Biotin was induced by addition of 1 mM CuSO4 to the medium for 16 h. a Heatmaps and profile plots of dBRD4-S-Biotin ChIP-seq in control (GST RNAi), NSL1 RNAi, MOF RNAi and WT untagged cells. The upper cluster comprises 2491 dBRD4-S-Biotin-bound and endogenous dBRD4-bound genes, the lower cluster of a similar number of randomly selected, not dBRD4-S-Biotin-bound genes. Both clusters are ordered by average intensity of all four ChIP datasets. Input-subtracted ChIP-seq data (merge of two biological replicates) is shown. b Boxplot of dBRD4-S-Biotin ChIP signal (log2 fold change over input) (merge of two biological replicates) on promoters of dBRD4-S-Biotin-bound genes (n = 2491) (±200bp from TSS) for the respective RNAi experiment. Boxplots show median (centre), interquartile-range (box) and minima/maxima (whiskers). Two-sided Welch two sample t-test was applied. c Representative IGV browser snapshots of dBRD4-S-Biotin ChIP-seq profiles in control (GST), NSL1 and MOF RNAi as well as in WT untagged cells, input-subtracted ChIP-seq data (merge of two biological replicates) is shown. d dBRD4-S-Biotin ChIP-qPCR in control (GST) and NSL1 RNAi as well as in WT cells serving as untagged control. Primers target the promoters and ends of dBRD4-bound genes. e NSL1 ChIP-qPCR of cells treated with JQ1 (5 µM) or DMSO for 4 h. f NSL1 ChIP-qPCR of cells treated with dBET6 (100 nM) or DMSO for 4 h. g NSL1 ChIP-qPCR of cells after control (GST) or dBRD4 RNAi treatment. For d–g data are presented as mean values, for d, e n = 2 or 3 biological replicates, for f, g n = 2 biological replicates. e–g Primers target the promoters and ends of NSL complex and dBRD4-bound genes, for expression changes in 4 h JQ1 treatment of these genes, see Supplementary Fig. 4f. Source data for d–g are provided as a Source Data file.